All Issues
Conventional and molecular assays aid diagnosis of crop diseases and fungicide resistance
Publication Information
California Agriculture 59(2):115-123. https://doi.org/10.3733/ca.v059n02p115
Published April 01, 2005
PDF | Citation | Permissions
Abstract
For the past decade, we have been developing techniques for monitoring pre- and postharvest diseases of tree fruit, nuts and vines at the UC Kearney Research and Extension Center (KREC). We have also advanced new methods to monitor pathogen resistance to fungicides, which growers can now use to make decisions on disease management. Although accurate, the conventional techniques are time-consuming and only provide results after 5 to 21 days. Molecular methods offer the possibility of faster, more reliable and efficient tests. We have developed such methods to monitor, diagnose and quantify crop pathogens. We have also used these new molecular techniques to answer complex questions on the biology of tree fruit and nut pathogens, the origin of their inoculum sources, changes in their population structures and the epidemiology of the diseases they cause. We are now working on molecular techniques that could supplement the conventional ones we have developed for vine diseases. Private diagnostic laboratories have been using a number of these conventional techniques and plan to adopt some of the molecular ones in the near future.
Full text
Our research at the UC Kearney Research and Extension Center (KREC) seeks to expand epidemiological knowledge of fungal diseases of tree fruit, nuts and vines in order to predict disease risk during the growing season — at harvest and during postharvest storage — and to predict resistance to fungicides. Both conventional and molecular methods can provide powerful decision-making tools for growers, enabling them to predict the emergence of latent disease, identify the sources and structure of fungal populations, and detect the emergence of pesticide resistance in fungal populations. Although the conventional techniques can be accurate and less expensive, only minimal information concerning a few isolates becomes available 1 to 3 weeks after testing. This can be a critical period for growers, who must make disease management decisions such as when and how to spray fungicides and which resistance-management programs to employ in the field.
Kearney scientists, including post-doctoral associate Zhonghua Ma, above, are developing advanced diagnostic tools to more quickly and accurately predict pre- and postharvest diseases of tree fruits, nuts and vines. For instance, spore-trap tapes in the field capture tiny fragments of DNA from airborne fungal spores, which can be identified within hours in laboratory molecular assays, bottom, while conventional methods require a week or more.
Currently, molecular methodology requires costly, specialized reagents and enzymes and expensive special equipment (PCR and RT-PCR machines) and is two or three times more expensive than the conventional methodology. Presently, none of the specific molecular methods are available to the average grower due to prohibitive costs and requirements for specialized lab facilities, equipment and specific training. Nor can UC research laboratories run samples for growers on a fee-per-sample basis. However, when affordable, portable molecular instruments and simple protocols are developed, we expect the routine, accurate and quick diagnosis of many crop diseases to be possible, even in the field. Private diagnostic laboratories have been adopting molecular methods for detecting and identifying plant diseases, and we recommend that they adopt the methods described here.
‘Howard Sun’ plum fruit show quiescent (small black specks) and latent (invisible) infections by M. fructicola, to compare conventional and molecular diagnosis techniques.
For example, we have learned that latent infections are correlated with subsequent disease emergence in the field or postharvest. Although conventional methods can detect latent infections, they are based upon a 1- to 3-week process of subjecting infected tissues to tissue-damaging agents (such as paraquat) or conditions (freezing), followed by the incubation of samples. Additionally, there are many variations in the type, number, duration and sequence of these processes. Molecular techniques can provide the basis for the efficient, accurate and rapid detection of pathogen inoculum — and are already being used to supplement critical conventional methods.
Predicting disease risk
The dynamics of plant disease development include: (1) the absence or presence and quantity of the pathogen's inoculum, (2) the stage and susceptibility of the crop and (3) the environmental conditions that are conducive to disease. These three factors are continuously changing and make the development of accurate predictive models difficult, complex and time-consuming.
Although increasingly accurate weather predictions are available through the National Oceanic and Atmospheric Administration (NOAA), other aspects of the disease triangle require a large database of information. The pathogen's spore inocula can be quantified by trapping spores, or predicted based on disease incidence in previous seasons (historical disease data). The most susceptible stage of the crop can be determined experimentally by periodic inoculations with the pathogen.
In many plant diseases, latency is an important epidemiological stage. In latent infections, the host and parasite coexist for a period with minimal damage to the host; latency involves an asymptomatic parasitic phase that eventually gives rise to visible symptoms if conditions are favorable for disease development (Verhoeff 1974). If latent infections are at high levels and develop into active symptoms, then a disease epidemic can occur. Latent infections of grapes, kiwifruit, stone fruit and nut crops remain inactive or may not survive until these fruit start to mature and environmental conditions become favorable for disease development. In most cases, the incidence of latent infections correlates with disease incidence in the field at harvest of tree fruit and nut crops (Michailides et al. 2000) or with the incidence of decays that develop in storage (Michailides and Morgan 1996).
The management of tree fruit diseases depends on the level of disease itself, environmental conditions, effective cultural manipulations, knowledge of pathogens’ sensitivity to fungicides, and the proper selection and timing of fungicide sprays. If growers had access to timely information on the presence of disease, the structure of fungal populations and the presence of fungal resistance to fungicides, they could make correct decisions on resistance management programs to avoid failures of disease control in the field or a buildup of resistance in the pathogen's population.
Fig. 1. Linear regression between incidence of latent infections (ILI) and percentage of branches with fruit rot (PBFR) caused by M. fructicola on prune, detected by the ONFIT technique. Each dot represents an average value of multiple locations and inoculations.
Conventional techniques take time
The accurate identification and early detection of plant pathogens are the cornerstones of successful disease management. The morphological identification of plant pathogens is often difficult and time-consuming and requires extensive knowledge of taxonomy and experience in recognizing detailed fungal features.
Direct agar plating technique (DAPT)
This technique isolates fungal plant pathogens from symptomatic plant tissues. The agar medium commonly used for the DAPT in our laboratory is acidified potato dextrose agar (APDA) adjusted to an acid pH of 3.5 by amending 2.5 milliliter of lactic acid (25% strength) per liter of medium. The acid pH is inhibitory to the majority of bacteria, but allows most fungal pathogens to grow when the Petri plates are incubated at 68°F to 77°F (20°C to 25°C) for 4 to 7 days. The DAPT can also be used to isolate latent infections from asymptomatic fruits and nuts using parts of tissues and contact areas (Michailides and Morgan 1997).
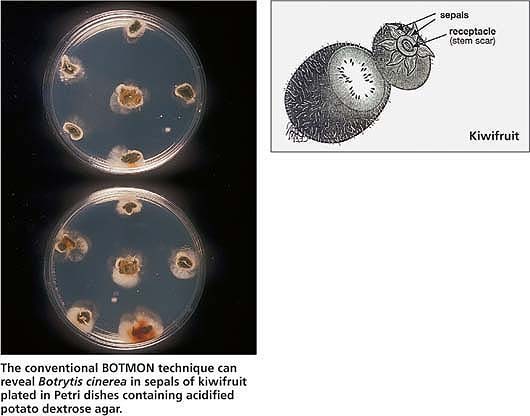
The conventional BOTMON technique can reveal Botrytis cinerea in sepals of kiwifruit plated in Petri dishes containing acidified potato dextrose agar.
This technique detects latent infections by Botrytis cinerea of kiwifruit (Actinidia deliciosa) sepals in the field. The incidence of latent infection is a good predictor of gray mold in cold storage (Michailides and Morgan 1996; Michailides and Elmer 2000). BOTMON involves the collection of fruit samples with stems attached, and the removal and plating of sepals or stem ends in APDA. We found that 1 month before harvest is the best time for sampling immature fruit to perform BOTMON and gray mold prediction. The results of the colonization of sepals and/or stem ends by B. cinerea become available 9 days after plating, and growers can use standardized tables to predict the levels of gray mold in cold storage.
In California, kiwifruit growers are increasingly using the BOTMON technique to make decisions on the need for preharvest fungicide sprays. Also, packinghouse operators and shippers use the results to determine needs for sorting and repacking fruit, to minimize secondary spread of disease in storage and to plan for fruit marketing. If growers spray only when needed (only vineyards with high incidence of latent infection), they reduce cost, increase revenue and reduce environmental contamination. The only disadvantage of the technique is that it is time-consuming.
Bud monitoring of Botryosphaeria (BUDMON)In the last two decades, we have seen more diseases caused by Botryosphaeria dothidea. In pistachios (Pistacia vera), it caused severe epidemics in 1998 with major yield losses. B. dothidea produces a multitude of spores in flasklike structures (pycnidia) that overwinter on the tree. Rains during summer, fall and winter spread the pathogen spores from infected parts of the trees to newly formed buds. As soon as buds develop, B. dothidea can infest them in latent form, allowing them to appear healthy, or can infect and kill them (Michailides and Morgan 2004). We developed a technique based on DAPT, which is used to reveal latent infections caused by B. dothidea in floral and vegetative healthy-looking buds of pistachio (Michailides 1991). Results are available 5 to 7 days after plating and incubation.
Overnight freezing incubation technique (ONFIT)
This technique is frequently used in our laboratory to detect and quantify latent infections of Monilinia fructicola and Monilinia laxa, which cause brown rot in stone fruit (fig. 1); Botrytis cinerea in grapes, which causes bunch rot preharvest and gray mold postharvest; and B. dothidea and Alternaria species, which cause blight diseases in pistachio. The test takes advantage of the fact that killing the fruit and leaf tissues at a stage when they do not favor disease development triggers the development of latent infections into active disease symptoms or accelerates the growth of hidden plant-tissue colonists. Our laboratory developed the ONFIT technique as an alternative to the use of the herbicide paraquat, which triggers the same events. Freezing and incubation eliminate the use of this toxic substance. While this is an improvement, the test still requires up to 7 days (Luo and Michailides 2003), during which an orchard could incur significant damage.
Molecular assays quick, accurate
We have developed several molecular methods that can supplement and may someday replace the slower conventional techniques. The polymerase chain reaction (PCR), which revolutionized molecular biology when it was first described in 1985, has been used to identify pathogens and determine their population structures, taxonomy and classification. This sophisticated yet increasingly common technology can definitively determine, for example, whether a piece of plant includes the genetic material of a microbe. Additionally, in the last several years we have applied techniques to quantify pathogen DNA, and these can be very useful to predict disease risk when the relationship between quantities of pathogens’ DNA in latent infections and disease levels has been established. Many plant pathologists are working toward establishing these relationships (the quantity of pathogen DNA to disease levels), especially now when recent advanced technology can quantify the pathogens’ DNA in plant tissues with great accuracy. However, because disease levels depend on weather conditions, it takes several years to establish such relationships. These techniques could aid in estimating disease potential and levels of latent infections relevant to disease in the field or during postharvest storage.
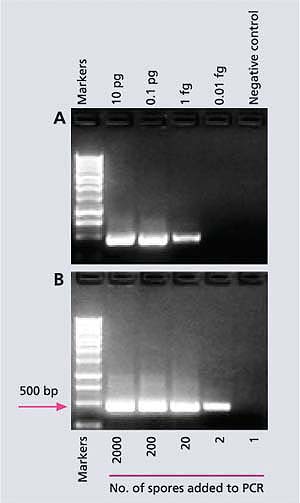
Fig. 2. (A) A sensitive molecular method (species-specific polymerase chain reaction) that can detect a fentogram (0.000000000000001 gram) of DNA from M. fructicola, the pathogen causing brown rot in stone fruit. (B) Another molecular technique (nested-PCR) can detect DNA of two spores of the pathogen. The tall column (gel) is a marker of the known molecular weight which is used as a reference to quantify the amounts of the short columns (gels). Base pairs (bp) is a unit showing the size of DNA; the larger the bp, the heavier (or longer) the DNA; pg = picogram (10-12 g); fg = fentogram (10-15 g).
Fig. 3. Molecular techniques can detect M. fructicola in (A) plum flowers collected from the field and (B) fruit inoculated with different concentrations of spore suspension of the brown rot pathogen and after various hours of incubation.
Brown rot in stone fruit
Brown rot, caused by M. fructicola or M. laxa, is a destructive disease of stone fruit (Prunus spp.) in California. The infected mummies of stone fruit harbor spores of brown rot pathogens, known as conidia and ascospores. Ascospores, produced only by M. fructicola, develop in cuplike structures (apothecia). (M. fructicola and M. laxa both produce conidia; however, conidia are not produced in apothecia.) The spores disperse in the air and infect blossoms, causing blossom blight. Subsequently, young fruit can become infected and eventually rot. When fruit bearing latent infections are thinned, they develop numerous conidia — especially under high relative humidity. Conidia can disperse and cause additional fruit infections in midseason. As fruit matures, a number of latent infections may develop into fruit rot. The inoculum potential (amount of a pathogen's spores) in orchards is an important factor in blossom blight and fruit infection (Luo and Michailides 2001, 2003). Determining inoculum potential in early- and midseason is critical for predicting brown rot accurately and managing it effectively.
However, inoculum potential is the most difficult disease parameter to determine in a stone fruit orchard. Currently, spore traps are used to determine the density of airborne disease agents, including M. fructicola. Because samples from traps require microscopic examination, this method is time-consuming and requires special training to recognize, count and culture spores, making it impractical for recording the large number of samples necessary for a large-scale disease management system.
PCR-based assays have the potential to monitor airborne inoculum levels of plant pathogens because they are highly specific and sensitive. Initially, we developed species-specific primers for the detection of M. fructicola in California stone fruits and flowers (Boehm et al. 2001). And recently, we developed a special PCR (nested-PCR), which is a very sensitive method for the detection of M. fructicola on sporetrap tapes (Ma, Luo, et al. 2003). First, sensitivity tests in the laboratory showed that the nested-PCR assay could detect the specific DNA piece in as little as 0.000000000000001 gram (or 10-15 gram) of M. fructicola DNA (fig. 2A) or in DNA from only two spores of M. fructicola (fig. 2B), which is a much higher degree of sensitivity than was previously possible. However, in a spore-trap tape sample from the field, the nested-PCR method can only detect 200 or more spores because of various inhibitors. Using these species-specific primers, we can also detect latent infections in fruit caused by M. fructicola within hours, while the DAPT or ONFIT would require at least 1 week (table 1). Since the nested-PCR assay cannot quantify the exact number of M. fructicola spores on a spore-trap tape, we are now working on a real-time (RT)-PCR technique that can do so.
Prune fruit show brown rot symptoms after processing with the overnight freezing incubation technique (ONFIT) and incubating for 7 days at 74°F (23°C).
Brown rot grows on prune fruit (without freezing) after sterilization and incubation with latent infections of M. fructicola for 2 to 3 weeks at 74°F (23°C) under 95% relative humidity.
Comparison of methods
In March and April 2001, hundreds of flowers of ‘Royal Diamond’ plum were collected from a commercial orchard in Reedley, Calif. A subsample of 30 flowers was divided into three groups based on visual symptoms: flowers heavily infected with M. fructicola and showing obvious signs of fungal sporulation on the stem and calyx surface, and designated (+); flowers displaying brown patches on the petals but no external signs of fungal sporulation (+/-); and flowers without any evidence of brown discoloration or fungal infection (-). Using these flowers, the PCR detected the DNA of M. fructicola in all (100%) of the (+) flowers, 80% of the (+/-) and only in 10% of the (-) flowers (fig. 3A).
These results were confirmed as accurate by plating (on APDA) another subsample of 30 flowers, providing a potentially useful method for assessing disease risk and blossom blight incidence and developing pre- and postharvest chemical control strategies against brown rot. This study took about 8 hours for collecting and testing the flowers. However, results from the plating of flowers became available after 4 days. Private laboratories could take this technology and offer it to growers right now. The PCR technique could supplement or replace the flower incubation technique (FIT), which provides an estimate of inoculum potential in stone fruit orchards in about 4 to 5 days.
In a second experiment conducted in an orchard with ‘Howard Sun’ plums, the presence of numerous visible quiescent infections suggested the presence of even more latent (invisible) brown rot infections. We decided to compare the DAPT of visible quiescent infections with the ONFIT of invisible latent infections and a species-specific PCR technique. In mid-May, fruit were observed in the field, and their fruit-to-fruit contact surfaces were marked with a permanent pen. All these fruit were then collected and brought to our KREC laboratory, surface-disinfected in 10% bleach solution for 3 minutes, rinsed with sterile water twice, and placed on clean paper towels. The fruit samples were split into three subsamples.
Using the PCR technique, 7.9% of the samples with invisible latent infections were positive for DNA of M. fructicola, and 6.7% of the fruit processed with ONFIT developed brown rot (table 1). Similarly, as expected when visible quiescent infections were used, 60.5% were positive for M. fructicola with the PCR technique and 54.3% of those plated on APDA developed colonies of M. fructicola. Most importantly, the traditional techniques required 5 to 9 days while the PCR technique provided the results within only 30 hours (table 1).
Pistachio fungi inoculum sources
As the pistachio industry in California has matured, fungal diseases have become a major threat, sometimes causing significant losses. One of these diseases, panicle and shoot blight caused by B. dothidea, was recorded for the first time in summer 1984 in a commercial orchard in Butte County. This disease infects shoots, branched flower clusters, leaves and panicles throughout the season. By 1999, it had spread to pistachio orchards throughout the state, causing yield losses of 40% to 100% (Michailides and Morgan 2004).
B. dothidea has been recovered from more than 35 native and introduced plant species (Michailides and Morgan 2004), suggesting that these hosts may serve as inoculum sources and threaten commercial plantings, since pistachio was only recently (early 1970s) introduced as a commercial crop in California. In greenhouse and field inoculation tests, we found that all B. dothidea isolates recovered from other hosts were capable of infecting pistachio.
Although multiple applications of the new strobilurin fungicides have provided excellent control of the disease in recent years, the potential development of fungicide resistance in B. dothidea is a significant concern. The use of resistant pistachio cultivars is the ultimate goal of any disease management program, but the judicious development and deployment of resistant cultivars depends on the population structure of the pathogen. To improve understanding of the pathogen's genetic diversity and how it has changed over the years at diverse locations, we conducted several studies that will eventually advance pistachio breeding programs (Ma et al. 2004).
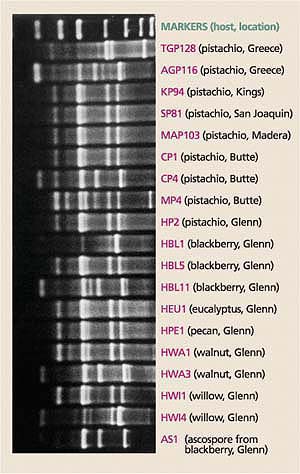
Fig. 4. DNA fingerprints of B. dothidea isolates (the pathogen causing panicle and shoot blight of pistachio) collected from hosts at different locations in California counties (shown in parentheses) and Greece.
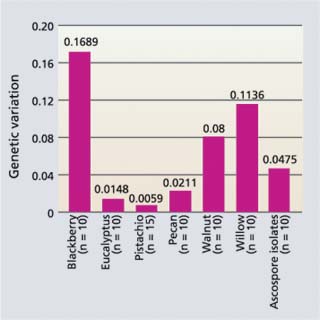
Fig. 5. Genetic variation within populations of B. dothidea from pistachio and other hosts next to a pistachio orchard in Glenn County. The taller the bar, the larger the genetic variation of isolates. Scale is from 0 to 1, where 0 = no variation and 1 = entirely (100%) different (1/5th of full scale is shown).
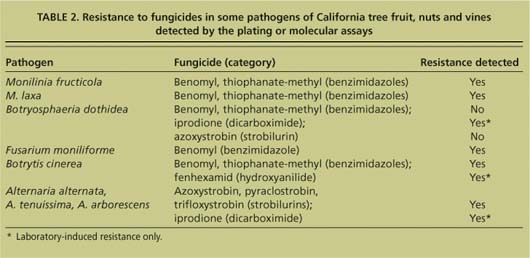
TABLE 2. Resistance to fungicides in some pathogens of California tree fruit, nuts and vines detected by the plating or molecular assays
Genetic variation
The genetic makeup of B. dothidea populations has remained homogeneous since it was first detected in 1984. We took samples of B. dothidea from infected pistachio orchards at diverse locations, some more than 600 miles (960 kilometers) apart, and over the course of several years. Our findings, as well as the pathogen's lack of propensity to form a sexual stage on this host, suggest that a program to breed resistant pistachio cultivars has a high chance of success. However, the interactions of a genetically uniform yet highly virulent pathogen population and the monoculture of the highly susceptible pistachio cultivars (‘Kerman’ and ‘Peters’) still pose a serious threat to the California pistachio industry.
DNA fingerprints
The DNA fingerprints of B. dothidea isolates collected from pistachio were identical to those of some B. dothidea isolates collected from other hosts (Ma et al. 2001), which suggests that native and introduced hosts for B. dothidea, other than pistachio, have historically served as potential inoculum sources for panicle and shoot blight of pistachio (fig. 4). (Forensic science uses similar DNA fingerprinting techniques in criminal investigations.)
Nonpistachio hosts
Although B. dothidea obtained from California pistachio are primarily asexual and very homogeneous, relatively high levels of genetic diversity were observed in B. dothidea isolates collected from nonpistachio hosts (fig. 5). These can infect pistachio. Consequently, if we are to select durable disease-resistant cultivars these cultivars should be tested with B. dothidea isolates that represent various genotypes from both pistachio and nonpistachio hosts.
Monitoring fungicide resistance
Fungicides are commonly used to manage plant diseases. However, the frequent use of fungicides with a single mode of action incurs a high risk of selecting resistant genotypes of plant pathogens. To determine levels of resistance to fungicides, the most common traditional technique is direct-plating single-spore isolates in media amended with various concentrations of fungicides and determining inhibition of growth and/or spore germination (table 2). The entire test can take 1 to 3 weeks — longer if the time required to isolate the pathogen from infected plant tissues is included. This conventional technique is time-consuming but critical, since growers rely on the results to decide on fungicide programs for their orchards.
In 2004, a new technique was reported using the spiral plate gradient-dilution method. Though an improvement, the method still requires 1 to 5 days for mycelial growth assays, followed by 14 to 20 hours for the spore inhibition studies to reveal results, plus 2 to 5 days for sporulation of fungi in culture (Förster et al. 2004).
DMA assays for fungal resistance
Strobilurin resistance
Alternaria late blight caused by Alternaria spp. in the alternata, tenuissima and arborescens species-groups (Pryor and Michailides 2002) is one of the most common fungal diseases of pistachio in California, and affects foliage and fruit. The disease can cause severe premature defoliation, staining of nutshells and molding of the kernels, which reduce fruit quality. Controlling Alternaria late blight requires a combination of cultural practices (irrigation management and pruning to reduce humidity) and multiple fungicide sprays.
The strobilurin fungicide azoxystrobin (Abound) provided excellent efficacy against this pistachio disease for some years. However, azoxystrobin-resistant populations of Alternaria have been detected in a fungicide experimental orchard and commercial pistachio orchards after multiple sprays for only 3 to 4 years, which caused failures of disease control in the field (Ma, Felts, et al. 2003).
Because azoxystrobin-resistant Alternaria populations appeared rapidly in pistachio orchards, it became urgent to incorporate effective antiresistance strategies when this or other strobilurins (because of cross-resistance) were used to control Alternaria late blight. The first important step is determining the sensitivities of pathogen populations to fungicides. Using the current conventional method, one person can examine only about 30 isolates in 2 work days, assuming that mature Alternaria spores are available for plating in fungicide-amended media. Obtaining spores requires 4 to 5 days for the incubation of cultures at optimum growth temperatures. It is almost impossible to produce results in a timely fashion.
Molecular markers by their potential nature are stable, not specific to the growth stage of the pathogen, and not influenced by the environment. If a DNA marker linkage with fungicide resistance is established (identification of the mechanism of resistance), it can be used to detect fungicide resistance in natural fungal populations.
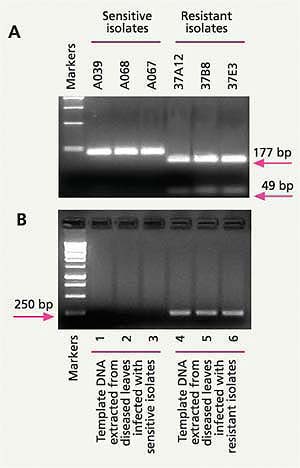
Fig. 6. (A) A change (mutation) in a specific gene of Alternaria spp. helps distinguish azoxystrobin-sensitive from azoxystrobin-resistant Alternaria isolates from pistachio; (B) A specific molecular technique (allele-specific PCR assay) can detect azoxystrobin-resistant Alternaria form lesions on pistachio leaves infected only by azoxystrobin-resistant and not by azoxystrobin-sensitive Alternaria isolates.
Surface sterilization of immature prune fruit with latent infections by M. fructicola is conducted by UC San Diego undergraduate Bobby Koja, by placing it in a container for the overnight freezing incubation technique (ONFIT).
In 2003, we found that azoxystrobin resistance in Alternaria from pistachio was correlated with a single-point mutation in the mitochondrial cytochrome b gene (Ma, Felts et al. 2003). Based on this mutation, we developed a PCR restriction fragment length polymorphism (PCR-RFLP) marker that detects DNA only from azoxystrobin-resistant isolates of Alternaria, but not from azoxystrobin-sensitive isolates (fig. 6A). Using this molecular method, one person can examine as many as 100 samples in a workday. Because many samples can be processed in a day, the cost for this test is not prohibitive. In fact, it is ready to be offered by private diagnostic laboratories that are willing to adopt and offer this new procedure (based on protocols provided by our UC laboratory).
Since the PCR-RFLP assay requires two steps and is still time-consuming if a larger sample must be tested, we further developed a one-step, allele-specific PCR (AS-PCR) method to rapidly detect azoxystrobin-resistant Alternaria in pistachios (Ma and Michailides 2004a). This AS-PCR method is being used in our laboratory now to rapidly detect the percentage of lesions caused by azoxystrobin-resistant Alternaria, making the results available to the grower within a day.
The AS-PCR assay was not quantitative per se and only detected the presence of azoxystrobin-resistant Alternaria. However, after purchasing a real time (RT)-PCR machine, the new RT-PCR technique can be used to accurately determine the frequency of azoxystrobin-resistant Alternaria isolates. Because 96 samples can be run simultaneously by the RT-PCR machine, the method is not expensive and is ready to be used by private diagnostic laboratories. However, a private lab has to first buy the expensive RT-PCR machine (about $35,000). Larger laboratories are equipped with RT-PCR machines, and we expect that within a few years smaller laboratories will adopt this technique.
Above, apothecia of M. fructicola as observed in the field and right, blossom blight in prunes caused by M. fructicola. New molecular technology can help growers to detect and better manage resistance to fungicides.
Based on the AS-PCR assay described above, we developed an allele-specific, real-time PCR assay to quantify the amount of azoxystrobin-resistant Alternaria DNA in a sample extracted from disease lesions (such as 50 disease lesions collected from a commercial pistachio orchard)(Ma and Michailides 2004b). Using this method, we can now detect frequencies of azoxystrobin-resistant Alternaria populations from at least 60 orchards (50 disease lesions per orchard) in 2 work days, while the conventional spore germination technique would require at least 200 work days for processing the same number of samples/isolates.
Benzimidazole resistance
Since benzimidazole resistance in M. fructicola and M. laxa has been shown to be genetically related to point mutations in the ß-tubulin gene (Ma, Yoshimura, et al. 2003), we developed an allele-specific RT-PCR method for rapidly detecting benzimidazole-resistant M. fructicola in stone fruit and M. laxa isolates in almonds, in a manner similar to that used for the detection of Alternaria resistance to azoxystrobin. This technique used blossoms collected from peaches and almonds in March 2004 and successfully detected the level of resistance to benzimidazole in M. fructicola (peaches) and in M. laxa (almonds). These protocols are available to private diagnostic laboratories, which are free to use the techniques at any time. Obviously, such rapid and quantitative detections of fungicide resistance in the fungal pathogen populations will be valuable for California growers to manage fungicide resistance in stone fruit and almond orchards, especially since the benzimidazole fungicide thiophanate methyl (Topsin-M 70WP) is still registered and recommended by the UC disease management guidelines to control brown rot diseases in stone fruit and almonds.
Future prospects
In general, molecular technology increases understanding of the biology and population structures of plant pathogens, provides quick and accurate answers to epidemiological questions about plant diseases, speeds up fungicide-resistance detection, and supports disease-management decisions. For example, in summer 2002 a pistachio grower in Kern County noticed that azoxystrobin had failed to control Alternaria late blight in his fields. Within 2 days of collecting Alternaria isolates from leaf lesions in his field, we cautioned that 80% of the Alternaria isolates were resistant to azoxystrobin and cross-resistant to other registered strobilurins. The grower avoided using any additional strobilurin sprays and instead he used chlorothalonil, which has a wide mode of action.
Future goals of our research are to develop techniques that can supplement or replace DAPT, BOTMON, BUDMON, FIT (flower incubation technique) and the conventional quantification of pathogen inoculum using spore traps in orchards and vineyards, with efficient, accurate and rapid molecular procedures using RT-PCR. In the near future, we plan to emphasize research on molecular techniques to provide results more quickly than the BOTMON and BUDMON techniques, especially since we have good evidence of the quantitative relationships of BOTMON with gray mold in cold storage and BUDMON with panicle and shoot blight of pistachio in the field. Furthermore, the goal of our laboratory is to reduce the cost of protocols that incorporate molecular techniques, by developing the capacity to process large numbers of samples at our laboratory and providing the protocols to private laboratories.
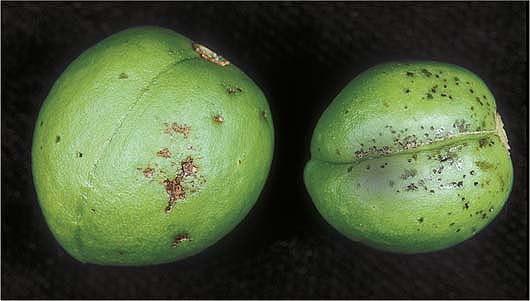
Right, symptoms of severe Alternaria late blight in pistachio; below right, leaf lesions caused by Alternaria isolates are used to determine the incidence of azoxystrobin-resistant isolates of Alternaria species. Below, pistachios severely infected with panicle and shoot blight caused by Botryosphaeria dothidea; DNA fingerprints of the fungus collected from pistachio and other hosts suggest that nonpistachio hosts serve as inoculum for the disease.