All Issues
Microbiome interactions and their ecological implications at the Salton Sea
Publication Information
California Agriculture 76(1):16-26. https://doi.org/10.3733/ca.2022a0002
Published online April 22, 2022
PDF | Citation | Permissions
NALT Keywords
Abstract
Although the Salton Sea was once a thriving destination for humans and wildlife, it has now degraded to the point of ecosystem collapse. Increases in local dust emissions have introduced aeolian (wind-blown) microorganisms that travel, along with contaminants and minerals, into the atmosphere, detrimentally impacting inhabitants of the region. Proliferation of certain microbial groups in regions of the Sea may have a disproportionate impact on local ecological systems. Yet, little is known about how the biogeochemical processes of this drying lakebed influence microbial community composition and dispersal. To elucidate how these microorganisms contribute, and adapt, to the Sea's volatile conditions, we synthesize research on three niche-specific microbiomes — exposed lakebed (playa), the Sea, and aeolian — and highlight modern molecular techniques, such as metagenomics, coupled with physical science methodologies, including transport modeling, to predict how the drying lakebed will affect microbial processes. We argue that an explicit consideration of microbial groups within this system is needed to provide vital information about the distribution and functional roles of ecologically pertinent microbial groups. Such knowledge could help inform regulatory measures aimed at restoring the health of the Sea's human and ecological systems.
Full text
As impacts of climate change and pollution worsen in the Salton Sea region, there is an urgent need to predict the Sea's ecosystem health and stability in response to water influx changes (e.g., increased evaporation, reduced inflow, and agricultural runoff irregularities). Although there is increasing evidence about the harmful effects of environmental degradation on wildlife within the Salton Sea, less is known about how microorganisms respond to mounting degradation throughout the region (California Natural Resources Agency 2020; Jones and Fleck 2020; Kjelland and Swannack 2018; Marti-Cardona et al. 2008; Moreau et al. 2007). Microorganisms and their respective communities (i.e., microbiomes) are ubiquitous, and the foundation of nutrient cycling within ecosystems (Falkowski et al. 2008). Research indicates that microorganisms are sensitive to natural and anthropogenic perturbations, and thus may serve as useful indicators of ecosystem productivity (Karimi et al. 2017; Maltz et al. 2017).
Wetlands in the Sonny Bono Salton Sea National Wildlife Refuge. To better understand the Salton Sea ecosystem, more research is needed on the interaction between the playa, sea and aeolian microbiomes. Photo: Jonathan Nye.
Without an explicit consideration of environmental microorganisms and their stress-response tactics, we may undermine our ability to respond to changing environmental and regulatory measures. For instance, policy decisions, plummeting water quality, and reduced Sea levels may have varying effects on microorganisms within this novel and vulnerable ecosystem. Moreover, understanding how the degraded environment surrounding the Salton Sea influences microbial processes, interactions and biogeochemical cycling is particularly important for assessing microbial contributions to overall ecosystem functionality, as well as for illuminating connections between policy- and climate-driven environmental changes and the health of nearby human and ecological systems.
Agricultural runoff, dust emissions
The implementation of the Quantification Settlement Agreement (QSA) in 2003 diverted water from the Colorado River to other areas, which massively reduced inflows to the Salton Sea (California Natural Resources Agency 2020). The New, Alamo and Whitewater rivers feed the Salton Sea with agricultural runoff containing pesticides, metals, salts and other elements (Vogl and Henry 2002). Specifically, copper, arsenic, manganese and selenium have been detected at levels above the U.S. Environmental Protection Agency threshold in water and sediment samples (Moreau et al. 2007; Xu et al. 2016). Selenium is of particular concern due to its consistently high concentrations in local fish (i.e., tilapia), at levels surpassing Aquatic Life Criteria standards (Xu et al. 2016). Federally banned pesticides including polychlorinated biphenyls (PCBs), dichlorodiphenyltrichloroethane (DDT) and dichlorodiphenylethane (DDE) have also been detected in in the muscle tissue of local fish species (Moreau et al. 2007; Riedel et al. 2002; Sapozhnikova et al. 2004; Xu et al. 2016) and in water, exposed lakebed (playa), and submerged playa samples (Sapozhnikova et al. 2004; Wang et al. 2012; Xu et al. 2016). Selenium, DDT and other pollutants accumulate in detritus, and are introduced into the Salton Sea's trophic network when consumed by algae, invertebrates and fish (Saiki et al. 2012). While the accumulation of these contaminants are detrimental to animal biodiversity (Canton and Van Derveer 1997; Köhler and Triebskorn 2013; Riedel et al. 2002), the extent to which pollution alters the Sea's trophic structure warrants further study.
In addition to pollutants, agricultural effluent delivers excess nutrients to the Salton Sea, leading to eutrophication and subsequent die-offs of aerobic organisms (Beman et al. 2005; Chaffin and Bridgeman 2014; Heisler et al. 2008). Nutrient enrichment leads to harmful algal blooms as these algae consume a majority of the Sea's dissolved oxygen. Eventually, these algae die off in the absence of sufficient oxygen, as other microorganisms decompose detritus and deplete the remaining dissolved oxygen (Qin et al. 2013). Additionally, strong winds during spring and summer seasons create upwellings of anoxic water and sulfide from the lake bottom to the surface (Marti-Cardona et al. 2008; Reese et al. 2008).
Algal blooms and gypsum blooms coupled with persisting anoxic conditions contribute to ongoing loss of wildlife, with particularly high mortality in local and migratory birds such as eared grebes (Podiceps nigricollis), fish such as Mozambique tilapia (Oreochromis mossambicus) and invertebrates such as pileworms (Neanthes succinea;Anderson et al. 2007; Carmichael and Li 2006; Marti-Cardona et al. 2008). Microbial pathogens, including Pasteurella multocida, cyanotoxins and botulinum toxin, have all been associated with mass die-off events (Carmichael and Li 2006; Meteyer et al. 2004; Nol et al. 2004); however, not all die-off events have been linked to heightened concentrations of these particular pathogens or toxins.
As required by the QSA, the volume of inflow will be reduced by 40% over the next decade, and the volume of the Sea itself will be reduced by more than 60% (Cohen 2014). Ongoing shrinkage of the Salton Sea not only increases salinity in the Sea and the playa (California Natural Resources Agency 2020), but also exposes additional lakebed sediment, leading to heightened dust emissions — dust production, wind erosion of sediment, etc. — in the area (Frie et al. 2017). These emissions are expected to contribute to already high levels of background particulate matter (Frie et al. 2017; Frie et al. 2019; US Fish and Wildlife 2014). Wet playas like the Salton Sea are vulnerable to erosion as capillary action in the sediment brings groundwater to the playa surface (Buck et al. 2011), softening sediment and stimulating groundwater evaporation (Reynolds et al. 2007). Playa emissions and aerosolized Sea spray contribute to the composition of the dust, which consists of minerals (e.g., selenium, sodium and sulfate), metals (e.g., cadmium and chromium; Buck et al. 2011; Frie et al. 2017; Frie et al. 2019) and dust-associated microorganisms, along with their respective microbial metabolites. These contributed materials, particulate matter size, and strong winds collectively influence dust composition, reduce local air quality and threaten downwind niches upon deposition.
Dust emissions disperse microbial components from dust to surrounding locations (Frie et al. 2017; Griffin 2007), and the harsh, arid climate of the Salton Sea provides habitat for microorganisms acclimated to these inhospitable conditions (Paul and Mormile 2017). Salinity, nutrient availability, pH, oxygen concentration and temperature collectively affect microbial composition and functional trait diversity. Most microorganisms are able to regenerate rapidly and transfer genes horizontally, which permits uptake of DNA from the environment as well as the sharing of DNA with other microbial species or viruses (Johnston et al. 2014). Altogether, these abilities allow microorganisms to easily adapt to the unique selective pressures of their environment.
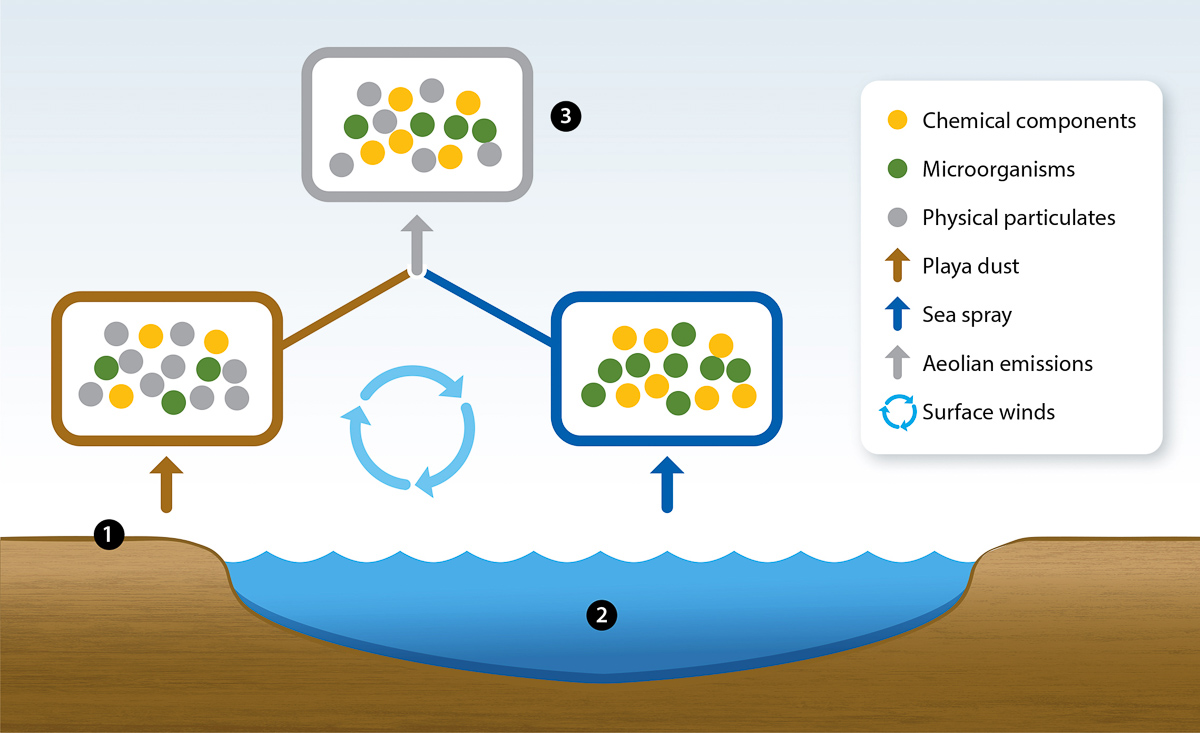
Microbial metabolic functions may be more dependent on environmental pressures than on evolutionary or phylogenetic relationships (Allison and Martiny 2008; Louca et al. 2017; Shade et al. 2012). Likewise, pollutants, excess nutrients and geophysical processes may alter the collection of microorganisms found within the Salton Sea's sub-ecosystems (fig. 1). The sub-ecosystem microbiomes include the playa, seawater and the wind-driven microorganisms that travel along with dust throughout the atmosphere (i.e., the aeolian microbiome). Interactions between environmental microbiomes and the ecosystem regulate the availability and accumulation of certain minerals. For example, anaerobic microorganisms in extreme environments akin to the Salton Sea can use selenate (SeO42-) or selenite (SeO32-) as electron acceptors; this reduces it to elemental selenium (Nancharaiah and Lens 2015), which accumulates in sediment. Other anaerobes can reduce sulfate to hydrogen sulfide, which consumes dissolved oxygen and yields harmful gypsum blooms, which are somewhat analogous to algal blooms (Ma et al. 2020). Clarifying how these microbiomes contribute to the trophic structures and chemical cycling within the Salton Sea is crucial to promoting the long-term sustainability and functionality of this ecosystem.
FIG. 1. Interactions among Salton Sea's dynamic sub-ecosystems. There are three environmental microbiomes: (1) playa, (2) seawater and (3) aeolian; sea spray and playa dust contribute to the aeolian microbiome. As the Salton Sea recedes, lakebed sediment is exposed and concurrently transforms into playa. Playa emits loose particulates that entrain microorganisms, chemicals and sediment into the atmosphere, which travel throughout the region via surface winds.
Playa microbiome
Because of their intimate associations within the Salton Sea, the biogeochemical interactions of the playa and the sediment beneath the lake are challenging to differentiate. Similar to microorganisms in Salton Sea water, anaerobes and extremophiles have a selective advantage in this niche due to the extremely high concentration of sulfate and salt in the sediments, which is compounded by the lack of oxygen and phosphorus resources (Swan et al. 2007).
Sediment depth gradients have been shown to differentially structure microbial communities. Most Archaea have been observed with equal abundance across lakebed sediment depths (Swan et al. 2010). However, Crenarchaeaota (i.e., Archaea phylum) and bacterial communities consistently exhibit similar abundances with depth. Likewise, the relative abundance of certain bacterial classes, including Betaproteobacteria, Gammaproteobacteria and Clostridia, correspond with both increased depth and salinity in the Salton Sea.
Several taxa found in Salton Sea playa have also been identified in marine sediments (Dillon et al. 2009; Swan et al. 2010) as well as haloalkaline lake sediments and salt flats (McGonigle et al. 2019; Rojas et al. 2018; Yang et al. 2016). Saline concentration has been identified as one of the most important factors in structuring microbial communities across ecosystem types (Lozupone and Knight 2007). These findings indicate that salinity and oxygen availability are crucial environmental drivers of microbial assembly in the Salton Sea playa.
Although the microbial composition beneath playa crusts has been studied to some extent (Dillon et al. 2009; Swan et al. 2010), the microorganisms of the superficial playa have largely been neglected. Increased playa exposure directly corresponds to greater dust fluxes in the region (Buck et al. 2011; Frie et al. 2019; Parajuli and Zender 2018), entraining both chemical and microbial components into dispersing dust (fig. 1). Heightened playa emissions correspond to salt precipitation on the playa surface (Buck et al. 2011), driving the microbial community structure at this playa-dust interface. Considering that playa surfaces are global dust contributors (Abuduwaili et al. 2010; Kandakji et al. 2020; Reheis et al. 2002; Reynolds et al. 2007; Ziyaee et al. 2018), characterizing and quantifying the impact of dispersing playa microorganisms on surrounding ecosystems and inhabitants of the region may be particularly important. Moreover, integrating these putative impacts into our understanding of wind-driven playa erosion may greatly advance our assessment of the vulnerability and toxicity of playa particulate matter. To understand the influence of both sediment and playa on dust composition — as well as associated exposure and deposition risks for downstream niches — the phylogenetic structure, functional diversity and seasonal variation of the playa microbiome must be investigated.
Sea microbiome
For decades, studies on the microbial composition of the Salton Sea have focused heavily on Cyanobacteria. Cyanotoxins — specifically microcystin — contribute to the high frequency of avian mortality events occurring at the Sea (Carmichael and Li 2006; Meteyer et al. 2004). Additionally, Cyanobacteria and other phytoplankton taxa form microbial mats that sit above the water surface (Wood et al. 2002) and thus are easy to investigate. Nevertheless, these studies have failed to capture the microbial diversity that persists below the Sea surface.
Microbial mat accumulation on the margin of Obsidian Butte during the warm summer months. Photo: Caroline Hung.
The Salton Sea is characterized by hypersaline, alkaline and anoxic conditions where anaerobes and extremophiles (i.e., microorganisms that inhabit extreme environments) prevail (González et al. 1998; Reese et al. 2008). These extremophiles include halophiles, or salt-loving microorganisms, and alkaliphiles, which thrive in conditions with a pH of 8 or greater (Andrei et al. 2012; Mesbah and Wiegel 2008).
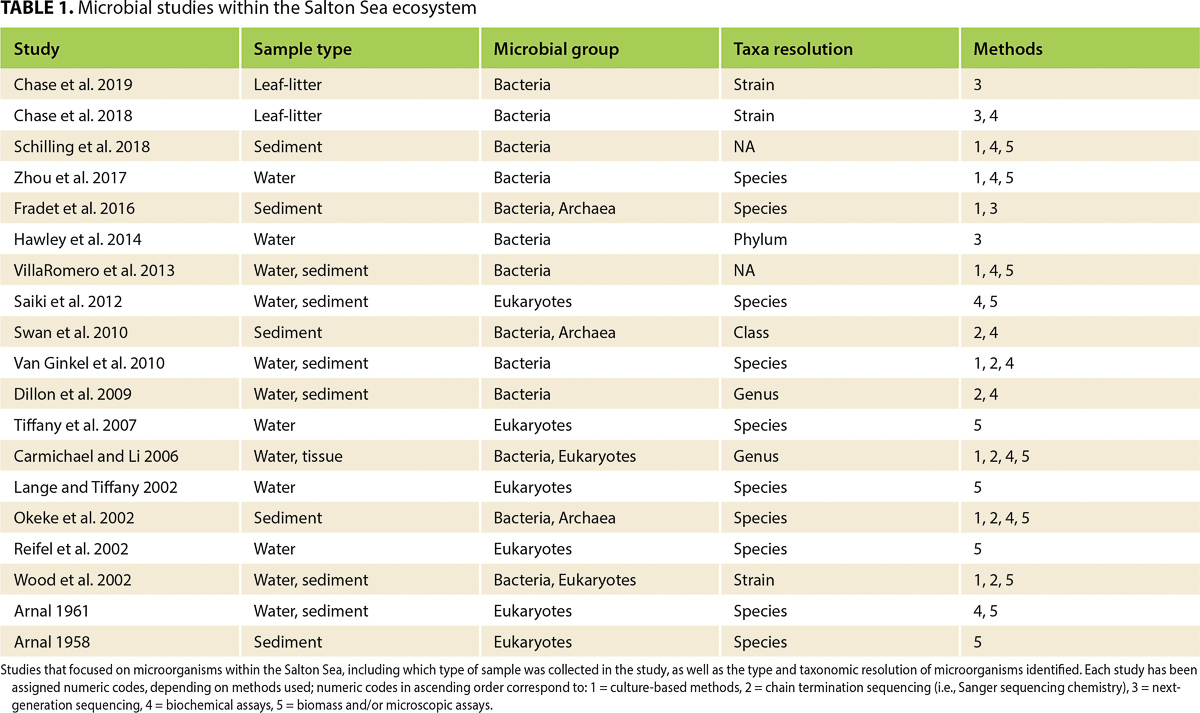
To date, only two studies have examined microbial phylogenetic diversity in Salton Sea water (Dillon et al. 2009; Hawley et al. 2014; table 1). In contrast to previous work, these studies showed that Cyanobacteria compose less than 5% of the total taxa found in Sea water samples (Dillon et al. 2009; Hawley et al. 2014); instead, they detected high abundances of microorganisms from both Proteobacteria and Bacteroidetes phyla. Beyond Cyanobacteria, Dillon et al. (2009) observed seasonal shifts in the relative abundance of Gammaproteobacteria and Alphaproteobacteria classes, and the Bacteroidetes phylum.
Proteobacteria are likely the most abundant phylum detected in Salton Sea water (Hawley et al. 2014). Rhodobacterales, an order within Alphaproteobacteria, was highly abundant within the summer months (Dillon et al. 2009). Rhodobacterales is composed primarily of photoautotrophs, which are capable of anaerobic photosynthesis, and haloalkaliphiles, which have been identified globally in saline, alkaline lakes (Kopejtka et al. 2017). Further determining which taxa within the Proteobacteria are most common and abundant would be a promising area for future research, as finer taxonomic resolution from this abundant phylum has rarely been reported. Given that a majority of these analyses about Salton Sea water capture microbial diversity only at the Sea surface, and neglect to characterize the microbial diversity of the water column, future work assessing microbiome structure below the water surface would be particularly valuable. For example, exploring the taxonomic diversity of microorganisms within the Sea's water column, particularly during seasonal upwellings and eutrophication events, would identify microorganisms responsible for depleting dissolved oxygen supplies via detritus consumption or sulfate reduction (Reese et al. 2008); classifying these microorganisms could improve predictions of anoxic periods within the Salton Sea and similar ecosystems.
Aeolian microbiome
In addition to minerals and trace metals, microorganisms can become entrained in both playa dust and Sea spray, along with their microbial toxins, which may be capable of withstanding turbulent conditions and long-distance transport (fig. 1; Tang et al. 2017). Microorganisms persisting among and on dust compose the aeolian microbiome, which includes all bacteria, archaea, fungi and viruses that circulate in the atmosphere. Some fungal spores and bacteria are capable of surviving within — or atop — dust as single cells or filaments, moving freely or attaching to individual particles (Samake et al. 2017).
Dust microorganisms have adapted to a unique set of environmental stressors including wind stress, ultraviolet radiation, humidity, temperature and nutrient availability. These microorganisms are equipped with a particular combination of traits, such as melanin production (Grishkan 2011) or biofilm formation (Aalismail et al. 2019), which enables their survival within this inhospitable airborne environment (Grishkan 2011). Several studies have reported higher microbial abundance on large dust particles at higher temperatures or low relative humidity (Lighthart and Shaffer 1997; Polymenakou et al. 2008; Yamaguchi et al. 2012), which are common features of the Salton Basin. Increased microbial burden also often correlates with enriched organic matter and minerals in dust (Tang et al. 2017). Collectively, these results suggest that larger dust particles shelter, sustain and protect microorganisms to ensure their dispersion and survival.
Numerous microorganisms isolated from dust remain viable for long durations of time. Bacterial and fungal isolates from dust not only can be successfully grown in the laboratory environment (Maki et al. 2019; Yamaguchi et al. 2012), but also have been shown to be metabolically active while in transit (Tang et al. 2017). Additionally, airborne microorganisms can facilitate ice nucleation, promoting cloud formation and precipitation (Amato et al. 2015; Bowers et al. 2009; Failor et al. 2017; Gaston et al. 2017). Therefore, aeolian microorganisms may threaten downwind ecosystems by altering precipitation and temperature, or disturbing stable microbiomes upon deposition. Moreover, exposure risks from airborne pathogenic microorganisms originating in the Salton Sea, and associated playa, may yield deleterious consequences for plants and animals, as well as human populations. Construction and farm workers may be particularly vulnerable to increased inhalation risks and exposure to dust-associated microorganisms, based on their occupational hazards (Gorris et al. 2018).
Although the microorganisms inhabiting the seawater and sediment of the Salton Sea have been examined, the dust microbiome has yet to be characterized. Given their remarkable ability to withstand environmental stress in the airborne environment, the aeolian microbiome could be dominated by either dormant or stress-resistant microorganisms. This resistant aeolian microbiome within Salton Sea dust could contribute to the health impacts of air quality in the region, especially if microbial groups break dormancy or actively interact with plant, animal or human hosts upon deposition. Some microbial adaptations likely permit their survival in dust. For example, bacteria from the genus Bacillus, which have been identified in dust samples from across the globe (Tignat-Perrier et al. 2019), have the ability to form endospores, allowing them to survive harsh environments via dormancy (Nicholson et al. 2000). Upon deposition, these dormant and resistant microorganisms may perform vital ecosystem functions, or they may pose formidable threats to inhabitants of the region. Motile microorganisms exhibiting chemotaxis may also be uniquely suited to explore porous environments and establish in favorable niches upon deposition (Scheidweiler et al. 2020). Therefore, exploring the functional attributes and microbiome structure within dust surrounding the Salton Sea would clarify the contribution of the aeolian microbiome to either promoting ecosystem stability or exacerbating regional public health crises.
Methods for further study
Future examination of the Salton Sea must comprehensively explore the playa, seawater and aeolian microbiomes to characterize their structural similarities, as well as any differences among these communities. Temporal and compositional differences may influence the interactions between these microbiomes and their surrounding environment, as well as overall nutrient availability within the Salton Sea ecosystem. Common methodologies used in human and environmental microbiome research could be tailored to explore both the taxonomic and functional diversity of Salton Sea microbiomes.
Sampling and analysis strategies
Multiple sample types (e.g., dust, water, playa) should be collected, at a variety of time points, from a replicated set of diverse locations within and around the Sea. As sample collection procedures will differ between media types (soil cores, dust collection, water samples), technological advances, such as the use of drones, water skimmers or seawater samplers (Xing et al. 2017), could be advantageous. Other valuable approaches may use semi-permanent passive samplers (Aciego et al. 2017; Frie et al. 2019), portable sampling platforms (Docherty et al. 2018) or active samplers, which collect all airborne cells and spores using filters from a known air volume (Frie et al. 2017).
Sample processing procedures may include filtering dust suspensions and seawater, using sterile 0.2-µm filters to capture bacteria and other microorganisms on the filter, while allowing passage of water and other aqueous substances. From unfiltered suspensions, microbial biomass can be determined using flow cytometry (Schmidt et al. 2020) or phospholipid fatty acids (PLFA; Buyer and Sasser 2012).
Amplicon marker genes, such as the 16S rRNA gene in bacteria or the internal transcribed spacer (ITS) region of fungal rRNA (Knight et al. 2018; Nilsson et al. 2019), are selected for amplification or quantitative polymerase chain reactions (qPCR; Manter and Vivanco 2007) to determine the taxonomic diversity and the relative abundance of important microbial groups across samples. Amplicon marker gene sequencing is currently the most cost-effective, high-throughput next-generation sequencing (NGS) method for studying microbiome composition across ecosystems (Liu et al. 2020).
Microbiome composition may be altered by both seasonal and agricultural geochemical fluxes within the Salton Sea, which also may select for metabolic strategies employed by these microbiomes. Sampling campaigns across time points at the same locations will capture temporal and seasonal variation, as upwelling events in the summer are known to change surface water chemistry by increasing sulfide levels and reducing dissolved oxygen content (Reese et al. 2008; Watts et al. 2001). Organo-chloride pesticides (OCPs) accumulate within previously underwater sediments, and may subsequently volatilize or evaporate as polluted sediments are increasingly exposed. As these pesticide-laden sediments become entrained in dust, OCPs are likely transported throughout the region via increased wind speeds and storms (LeBlanc et al. 2002), exacerbating dust inhalation risks. Although microorganisms from the playa or aeolian microbiomes may be capable of metabolizing recalcitrant or labile components from polluted dust, ecophysiological assays and metabolic models would be required to quantify the extent to which these toxins can be transformed or biodegraded by extant microorganisms. Future work incorporating and altering model parameters would facilitate our ability to predict future fate and transport of toxic dust based on future water influx and climate change scenarios (D'Amato et al. 2008).
Changes in geochemistry, such as total organic carbon concentrations, pH and nutrient levels, have pointed to detectable shifts in playa microbiomes in similar hypersaline water bodies (Hollister et al. 2010). Collecting soil core samples from the playa over a distance transect would help to elaborate how geo-chemical variation impacts Salton Sea terrestrial microbiomes. Furthermore, as reductions in sea volume expand the exposed playa, these sampling strategies and subsequent analyses could clarify how microbiomes transition with their environments from moist to dry conditions.
Lake sediment cores sampled from the Salton Sea by the Lyons lab at UC Riverside for trace metal contamination analysis. Photo: Caroline Hung.
The impact of dust composition and evaporite minerals (e.g., magnesium, calcium, sulfate) on aeolian microbial metabolism and assembly could be studied via deploying dust collectors (Aarons et al. 2019; Frie et al. 2019). Dust samples can be analyzed using stable isotope ratios, such as 87Sr/86Sr and 143Nd/144Nd, to detail the provenance of the dust and its corresponding microbial community (fig. 1; Aciego et al. 2017; Dastrup et al. 2018; Xie et al. 2020; Yan et al. 2020). Furthermore, Sr-Nd isotopic analyses of Salton Sea dust may reveal geochemical features of the dust's provenance (i.e., the geography and climate) that may select for microbial migration from the Sea and the playa to the aeolian microbiome. Coupled with NGS technologies, this comprehensive analytical approach will explicate the dynamics within the playa, seawater and aeolian microbiomes, as well as their associated implications for microbial dispersal throughout the Salton Sea Basin.
Meta-Omics analyses
Exploring diversity in Salton Sea microbiomes could leverage sophisticated molecular techniques (i.e., -omics), such as high-throughput NGS methods, shotgun metagenomics, metatranscriptomics or metaproteomics. Briefly, a shotgun metagenome describes the collection of genomic material from a particular ecosystem, including both eukaryotic and prokaryotic genomes (Quince et al. 2017). Metagenomics can be used to characterize taxon-level microbial diversity and categorize putative functions performed by the microbial community. Although metagenomics can identify the functional traits within a microbiome, metatranscriptomics confirms which traits are actively expressed by the microbiome at a given time. A metatranscriptome includes the totality of gene (i.e., RNA) transcripts found within an environment (Shakya et al. 2019). Recently, metaproteomics has been used to complement NGS methods, via classifying and quantifying proteins produced by microbiomes (Hettich et al. 2013). Metagenomics has been used to study Salton Sea leaf litter (Chase et al. 2018; Chase et al. 2019; table 1) and seawater (Hawley et al. 2014), but these studies did not detail the functional diversity of their samples. Salton Sea playa and aeolian metagenomes have not been thoroughly described, nor have metatranscriptomic or metaproteomic approaches been employed yet for characterizing the activity of Salton Sea microbiomes. Because of the dearth of information on microbiome structure within the region, the Salton Sea presents a unique opportunity to utilize -omics techniques to study both microbial taxonomic and functional diversity across sub-ecosystems, which compose the larger ecosystem (fig. 1).
Metagenomic, metatranscriptomic and metaproteomic analyses have been performed on a wide variety of sample types, including the human gut (Long et al. 2020), soil (Romero-Olivares et al. 2019), deep-sea sediments (Mason et al. 2014), cloud water (Amato et al. 2019) and airborne dust particulates (Aalismail et al. 2019). These techniques allow for the comparative analyses of microbial genomes, transcriptomes and proteomes from different systems and ultimately identifies both shared taxa and genes among microbiomes. Functional annotation of metagenomes and metatranscriptomes, using comprehensive databases such as the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al. 2016; Kanehisa et al. 2017; Kanehisa et al. 2019) and analytical tools like KoFamScan (Aramaki et al. 2020), detail the functional traits that are differentially expressed under specific environmental conditions (Amato et al. 2019; Chung et al. 2020; Shakya et al. 2019). For instance, soil metatranscriptomes may indicate whether microbial communities are actively allocating resources to stress response or proliferation (Romero-Olivares et al. 2019). Functional annotations can then guide the classification of proteins, such as microbial exudates, identified in metaproteomes (Hettich et al. 2013). Collectively, these analyses may further reveal metabolic strategies that enable microbial persistence in harsh conditions (Brewer et al. 2019). Furthermore, understanding these associated metabolic processes may reveal mechanisms that drive microbiome-resource interactions throughout this dynamic ecosystem.
Modeling dust emissions
Mineral dust advection, or dust transference by fluid flow, has been shown to be an important vector for the long-range transport of microbial organisms, especially in and around desert environments such as the Salton Basin. While smaller particulates have longer atmospheric lifetimes due to their slower deposition velocities, larger aerosols (> 5 μm) typical of desert surfaces are especially efficient vectors of microbial dispersal (Yamaguchi et al. 2012). Dust transport patterns and ranges are thus dependent on both particle size and meteorology, with strong wind systems capable of relocating larger particles — along with any attached microorganisms — across continental distances (Perry et al. 1997). The Salton Sea region is one of the dustiest in the United States, with both the Coachella Valley and Imperial Valley regions consistently exceeding daily EPA standards for particulate matter (PM10). Moreover, these regions exhibit strong seasonality and frequent wind storm-driven dust events (Evan 2019; US Environmental Protection Agency n.d.). This makes the patterns of dust emissions and transport, as driven by seasonal meteorology and sporadic dust storms, important for understanding regional sources and biogeographic patterns of local microorganisms.
Available tools for identifying sources of advected dust include trajectory or dispersion models such as the Hybrid Single Particle Lagrangian Integrated Trajectory Model (HYSPLIT; Stein et al. 2015). By combining local wind fields with physical dust deposition parameters, these models can run forwards (to estimate patterns of dust transportation and deposition from a given source) or generate backwards trajectories (to assess likely emission sources for particles collected at a given receptor site). These methods have been used previously to examine long range transport patterns of source-specific microbial populations (Cáliz et al. 2018; Rosselli et al. 2015; Stres et al. 2013; Yamaguchi et al. 2012) and impacts of dust storms on downwind microbial communities (Hagh Doust et al. 2017; Mazar et al. 2016).
Our group has generated backward trajectories to evaluate the wind patterns blowing from the Salton Sea and estimate relative contributions of particulates reaching our passive dust collectors over a finite time period. Furthermore, we have explored the distribution and elemental composition in the Salton Sea region (Frie et al. 2019), which provides valuable context for explaining how seasonally shifting dust patterns — along with chemicals, physical particulates and associated microbial transport — may influence local microbiomes.
Conservation and public health in the Sea
Characterizing the unique microbial communities of the Salton Sea will complement ongoing investigations of the impacts of pollution on local residents and wildlife. Yet, many questions remain. For instance, how does the aeolian microbiome influence the lung microbiome? The aeolian microbiome may exacerbate respiratory symptoms via incidentally inhaled aeroallergens and particulate matter, resulting in the disproportionately higher rates of asthma and chronic respiratory disease detected in nearby communities (California Department of Public Health n.d.; Farzan et al. 2019). Can we explain long-term exposure effects of Salton Sea dust on the health of wildlife and local residents by comparing the Salton Sea microbial communities with unpolluted microbial communities, collected in analogous systems? The microorganisms themselves, in addition to their extracellular exudates (Chae et al. 2017; Rolph et al. 2018), may serve as bioindicators of either eutrophication or pollution in soil, water or dust (Bouchez et al. 2016; Karimi et al. 2017; Schloter et al. 2018). Furthermore, historical exposure to pollutants may select for microorganisms that are uniquely suited for tolerating — or even ameliorating — toxicity within these particular systems.
In the interest of augmenting restoration efforts, how can we best deploy particular microbial taxa from the Salton Sea sub-ecosystems to remediate polluted systems via biodegradation or metal transformation (i.e., bioremediation; Kumar et al. 2019; Sher and Rehman 2019; Voica et al. 2016)? Novel opportunities for restoration may arise as human activities, such as mining and food production, increase apace with the shrinking of the Salton Sea. To illustrate one example, lithium mining of geothermal brines in the Salton Sea (Vikström et al. 2013), coupled with evaporation, may provide opportunities to leverage endemic microorganisms for bioremediation. Yet, microbial bioremediation may not be sufficient to mitigate the environmental impacts and deleterious human health outcomes for inhabitants of the region exposed to air pollution; this pollution may be exacerbated by evaporating novel brines, replete with toxic metals such as arsenic and manganese, which may cause neurological issues in children (Dion et al. 2018). To promote community health and ecosystem stability, we must investigate the dynamic interactions among the playa, seawater and aeolian microbiomes throughout the region. Furthermore, a thorough characterization of the functional attributes of dust microbiomes is needed to inform holistic approaches for addressing regional public health crises throughout the Salton Sea Basin.
Conclusions
The Salton Sea crisis necessitates immediate action as conditions rapidly worsen. Reduced precipitation and increasing temperatures are drivers of drought throughout California (Luo et al. 2017), advancing the diminution of the Salton Sea. Playa erosion and resulting dust emissions are predicted to rise (Parajuli and Zender 2018), which could interfere with incoming radiation and induce subsequent changes to local climate (Von Schneidemesser et al. 2015). Fluctuations in nutrient availability as a result of climate shifts will select for specific microbial functions (Louca et al. 2017; Louca et al. 2018), altering the overall trophic structure in the Sea. To better understand ecosystem resilience in this unpredictable landscape, more research is needed on the functional potential of these interacting environmental microbiomes and their contributions to nutrient cycling.
Any actions taken to increase the stability or conservation of this ecosystem may have public health implications, and vice versa, and we must anticipate the consequences of inaction to humans and wildlife. Careful consideration of the impacts of restoration or mitigation attempts must be holistically examined, as approaches addressing one area of concern may inadvertently yield adverse consequences for other areas. With greater knowledge, resources can be allocated towards strategic measures that aim to ameliorate the health of local human populations and promote the restoration of diverse wildlife and microbial communities that support resilient ecosystems.