All Issues
Integrated data-collection system tracks beef cattle from conception to carcass
Publication Information
California Agriculture 64(2):94-100. https://doi.org/10.3733/ca.v064n02p94
Published April 01, 2010
PDF | Citation | Permissions
Abstract
Data on the performance of individual beef cattle from birth to processing can be used to improve herd management and genetic characteristics such as fertility, weaning weight and tenderness. Likewise, tracing the origins and whereabouts of cattle that may have been exposed to infectious disease agents is necessary to protect the health of the national beef herd. Such information is rarely available, in part because cattle often change ownership during the beef production process. A system developed to support the lifetime tracking of individual cattle in the UC Davis beef herd seamlessly transfers information between the UC Sierra Foothill Research and Extension Center, UC Davis campus feedlot and a commercial beef-cattle processing plant, and provides a repository for performance data collected at all production stages. The system provides real-time data sharing, as well as integrated analysis and management evaluation options, and will be a valuable resource for beef-cattle research. UC livestock farm advisors are now implementing a similar system with cooperating commercial ranches.
Full text
The beef-cattle supply chain consists of several distinct sectors. An animal may change ownership several times during its life, and each transfer presents a major obstacle for the dissemination of individual animal records from one sector to the next. Often animal health data and other important information are never shared among members of the supply chain.
A model cattle-identification system tracks animals from birth at the UC Sierra Foothill Research and Extension Center, to feedlot and harvest. Researchers are using the system to better understand cattle genetics, beef qualities and other characteristics.
The cycle typically starts in the cow-calf sector, where the animals are bred, born and raised. Once animals reach a certain age or weight, or their feed resources become limiting, they are sold, sometimes to a stocker operation for additional weight gain before they are sold again to a feedlot. At the feedlot, animals are “finished” — often fed on a grain-based diet until they reach market weight, at which time they are harvested. Carcasses are then transferred to the packer where they are divided into wholesale cuts, then into retail cuts.
In the absence of an integrated identification system, tracing a single steak back to its carcass, let alone back to the cow-calf operation where the animal originated, becomes extremely complicated. Although steak may be the end-product, it is not the end of the chain. That position is reserved for the consumer, and it is ultimately the consumer's eating experience that should be the driving force for selection and management decisions throughout the supply chain. However, because most cow-calf producers do not retain ownership of animals after they leave the ranch, they receive no information on (1) animal performance at the feedlot, (2) carcass quality attributes or (3) consumer satisfaction with products derived from the animals they bred and raised. Consequently, they receive no price premiums or discounts.
Industry fragmentation is also a major obstacle hindering the effective traceback of animal disease outbreaks. Animal diseases pose a major threat to livestock production in the United States, and the intentional or unintentional introduction of infectious disease agents could have a catastrophic impact on cattle producers. A 2006 study estimated that the combined consumer and producer losses from a foot-and-mouth disease outbreak in the United States would exceed $266 billion (Zhao et al. 2006).
Although approaches to enable traceback for investigation and response to disease incidents have been discussed nationwide, implementation has been slow. The U.S. Department of Agriculture (USDA) National Animal Identification System (NAIS) has met with considerable resistance for a variety of reasons, including uncertainty about proposed equipment and systems, and the absence of any direct benefit to the producers who must pay for its implementation. Currently, participation in NAIS is voluntary and limited. However, other beef-producing countries — including Australia, the European Union, Japan, Brazil, Argentina and Canada — have already adopted such systems, usually in conjunction with government subsidies to encourage participation, and they are enjoying the benefits of source and age-verified marketing opportunities. The end result is that U.S. beef producers are falling behind in technology adoption, and their responsiveness to biosecurity issues is limited. At the same time, the public may perceive that the beef industry is unresponsive to animal health and food safety issues.
To address these problems, a collaborative research team developed a fully integrated, electronic, individual-animal tracking-system prototype for the UC Davis Department of Animal Science cow-calf herd, which is used for research and education and is located at the UC Sierra Foothill Research and Extension Center (SFREC). Vertical integration of the system provides opportunities for researchers to analyze commercial beef-cattle production from conception to carcass. The overall objective of this project was to demonstrate that a functional, integrated data and trace back system could be useful for cattle management, while simultaneously providing biosecurity features. The project team included staff at SFREC, the UC Davis campus feedlot, employees of a commercial processing plant, UC farm advisors, and researchers from the UC Davis Department of Animal Science and School of Veterinary Medicine.
Round electronic ear tags, inset, were placed in the left ears of SFREC cattle and used as the key integrating identification number for cattle records. Visual ear tags, electronic wands and various electronic devices were also used to collect data.
Remote-access antennae (yellow ovals) were used to transmit data from cattle chutes to computers in the SFREC office.
System design
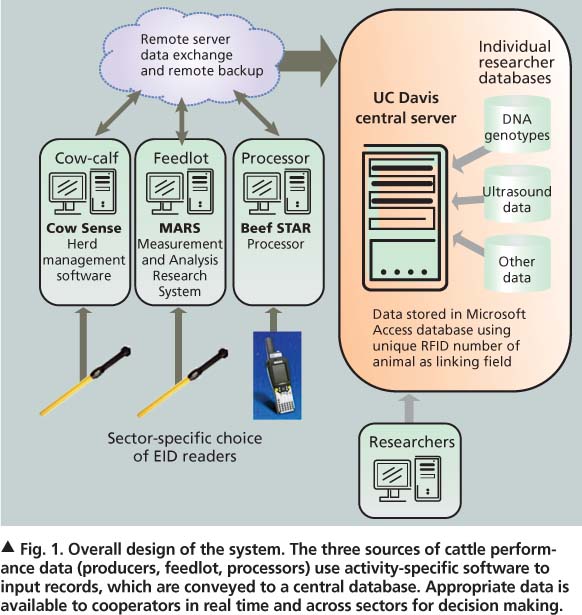
Conceptually, the data management system now in place for the UC Davis cow-calf herd comprises several distinct operations (fig. 1). Since 2007, various sectors (users) have been collecting data with commercially available software designed for their specific needs.
SFREC.
Data collected at SFREC includes calving date, birth weight, mother-of-calf identification, weaning weight and health records. All (approximately 250) calves are fitted with individually numbered ear tags at birth. The SFREC herd manager enters all records into Cow Sense herd management software (Midwest MicroSystems, Lincoln, NE). Subsequently, a computer running Cow Sense is used to collect data on location at the handling chute each time cattle are brought through. That information is then remotely transferred to the main office at SFREC where it can be updated before being linked through the Internet to the UC Davis central computer and stored in a Microsoft Access database.
Feedlot.
Data is available in real time when calves are shipped to the UC Davis feedlot. This information is valuable to feedlot operators when assigning calves to specific dietary, production or other experimental groups. Calves are weighed upon feedlot entry and exit, and at 30-day intervals to determine the average daily rate of gain. Feedlot data is collected using the Measurement and Analysis Research System (MARS) (Midwest MicroSystems), a computer program that allows for repeated measurements of multiple variables.
Harvest.
Cattle are harvested based on weight and visual estimates of finish, and shipped in groups of approximately 20 head. Carcass data is collected from a commercial harvest facility in Los Banos, Calif., by a USDA grader using a hand-held scanner (PSION Workabout Pro) similar to those used by overnight delivery services (fig. 1).
Database connections.
Data is delivered to the UC Davis server via software called Beef STAR (Midwest MicroSystems), with databases from the three sectors connected using Microsoft Access. Data also flows back to the herd management software, Cow Sense, to provide the SFREC herd manager with information on calf performance in the feedlot and at harvest. This feedback allows for the development of on-ranch genetic evaluations for feedlot performance and carcass traits, not typically an option for commercial cow-calf producers.
UC researchers also have linked ancillary databases of detailed research data to the central database. Alison Van Eenennaam developed a DNA database that contains SNP (single nucleotide polymorphism) genotypes for each bull, cow and calf at SFREC. Other UC researchers are beginning to tie into the system, including Bruce Hoar of the UC Davis School of Veterinary Medicine, who is the attending veterinarian for the UC cow-calf herd at SFREC. This collective information enhances the value of data from each individual researcher, and both the content and number of animals in the database will continue to grow over time.
Unique identification numbers
Data is collected from each calf crop from conception to carcass over a period spanning 2 to 3 years. At any point in time, three crops consisting of 100 to 250 calves are either being gestated, calved and grown at SFREC, or finished at the feedlot. It is easy for animal numbering systems to become confusing. Two animals with the same number (e.g., no. 1) might exist concurrently as a weanling at SFREC and a steer being fattened at the feedlot. The key to keeping accurate records is assigning a unique identification number to each animal. This is done using a radio-frequency-identification (RFID) ear tag transmitting an embedded 15-digit number, which is assigned to each animal in the cow-calf herd at SFREC. The small, round button tags are placed in the left ear and read by hand-held or chute-side electronic readers as the animal moves through the production process. The tags provide rapid and error-free recording and tracking of individual animals. The identification number links information about each animal, whether it concerns assigning the calf to DNA-based parentage determinations, weaning weight, feedlot gain or the eventual quality grade of its carcass or size of the rib-eye steak it produced.
At harvest, when carcass data is collected, reliable identification takes on additional importance. The detection of a contagious animal disease could potentially trigger an urgent need to determine the source and recent movements of the animal relating to a particular carcass. In the event of such a traceback, linkage to all previous production records from birth to harvest would be critical for a timely response. Knowledge of each animal's whereabouts and origin would facilitate a targeted health response, as opposed to a mass recall and potential depopulation of unaffected herds. This would increase response effectiveness and decrease the costs associated with an animal disease outbreak.
Some of the problems that we encountered during the development of this integrated data-collection system included implementing electronic identification technology in a practical manner for the field collection of data, database sharing structure, data integrity and security, the education of collaborators and staff working on the project, and the timeliness of data availability for real-time use. The system was designed to be dual purpose: first, to accomplish traceback in the case of a disease outbreak, and second, to develop valuable information for cattle management, thereby rewarding data collection by all parties.
Fig. 1. Overall design of the system. The three sources of cattle performance data (producers, feedlot, processors) use activity-specific software to input records, which are conveyed to a central database. Appropriate data is available to cooperators in real time and across sectors for decision making.
Fig. 2. Sire prolificacy data for 3 years (2006-2008) with sires present in multiple years. Each calving group had two to four bulls, and the number of cows per pasture varied from year to year. The first digit of each bull's identification number is the last digit of the bull's year of birth (i.e., bull 362 was born in 2003). Large variations in prolificacy year-to-year and among sires were observed, and prolificacy of a sire in the first year was not always predictive of subsequent prolificacy when sire-breeding groups changed.
Breeding performance
We were able to apply data obtained though the integrated data-collection system in a study of cattle paternity and breeding success. Commercial beef cattle herds using multiple-sire breeding pastures often have no way of identifying which bull fathered a calf, or which bull produced the best (or worst) cohort of calves. Inherited DNA markers can be used to assign paternity. Bulls pass on only one of the two copies of each gene or “marker allele” that they carry. Paternity identification involves examining each calf's genotype at multiple locations in the genome and excluding as potential sires those bulls that do not share common alleles with the calf (Van Eenennaam, Weaber, et al. 2007). DNA was extracted from tissue collected from all bulls housed at SFREC before they were turned out with the cow herd for the 2006, 2007 and 2008 breeding seasons, and DNA was collected from all calves delivered during those 3 years. All DNA samples were genotyped using a 99 SNP panel (Igenity, Duluth, GA), and the results were used to match potential sires to their offspring.
Sire performance in terms of the number of offspring produced was highly variable from year to year (fig. 2). For example, bull 357 was a sire all three years, but in the first year he sired only one calf, during the second year he improved slightly to sire six calves, and in the third year he sired nine calves, for 16 calves total. In contrast, while in 2006 bull 115 was only present for the first year, he sired 25 calves. At first glance, it would appear that bull 115 far outperformed bull 357; however, consideration must be given to the size of the breeding groups (or how many cows were available to be bred) and how many competing sires were in the group. Bull 357 was in a four-sire breeding group for the first two years (with two older, and thus likely more dominant, bulls), and a three-sire breeding group (with two older bulls) in the third year. This means he had to compete with older bulls in every breeding season. In contrast, bull 115 had only one young sire as competition the year that he was an active sire in this evaluation.
We have examined sire prolificacy (breeding success) on a number of commercial cow-calf herds over the past few years, and have consistently seen this variability in calf output (Van Eenennaam, Weaber, et al. 2007). Other studies have reported similar variability in calf output among herd sires, and further, found that prolificacy is moderately repeatable if the composition of bull mating groups remains similar (DeNise 1999; Holroyd et al. 2002). This information is important from the standpoint of genetic improvement because prolific bulls will have a greater impact on herd genetics. Reconciling DNA information on sires and calves is the only way to assign parentage in multiple-sire breeding pastures.
On-ranch genetic evaluations
Another area in which comprehensive animal tracking is invaluable is in improving cattle performance through selective breeding. Breeders can make genetic progress by determining the genetic potential of animals as parents. In the beef-cattle industry, an estimate of genetic potential or breeding value is called an expected progeny difference (EPD) (see box).
Graduate student Gustavo Cruz records ultrasonograms at the feedlot as part of research activities with the UC cattle herd.
It is particularly important to obtain accurate estimates of the genetic potential of bulls, as bulls will produce more offspring than cows during their lives. Genetic evaluations of young natural-service bulls are often based on the average genetic potential of the parents and observations of the bull itself, and these estimates cannot be further improved in the absence of progeny data. DNA markers cannot only resolve the paternity of offspring produced in multiple-sire breeding pastures, they can also be used to better estimate the genetic worth of natural-service bulls through on-ranch progeny testing (Pollak 2005).
Measuring bull performance
Expected progeny difference (EPD) is the difference between the average performance of a bull's progeny and the average of those sired by another bull. Breed associations develop the most commonly available EPDs based on their extensive nationwide databases of pedigree and performance information. In the absence of other information, the genetic merit of an animal can be predicted based on the average breeding value of its parents. This generates a low-accuracy “pedigree estimate” that is typically associated with young animals prior to the collection of any information on their own performance. With only ancestor information, full siblings will have the same EPD. Their true value will vary, however, as a result of the random inheritance of parental genes. Incorporating progeny performance information increases the accuracy of EPDs. This can be seen in beef-sire semen catalogs, where very-high-accuracy EPDs are associated with bulls with many progeny as a result of their use in artificial-insemination programs.
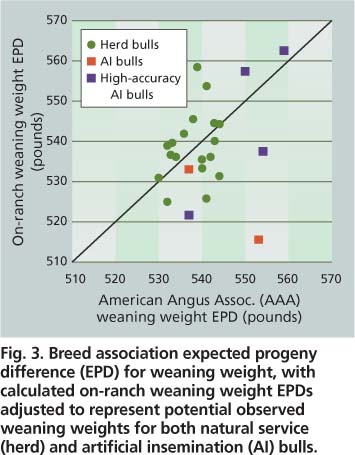
To illustrate these concepts, we compared on-ranch and breed-association EPDs for a set of 21 natural-service bulls and six artificial-insemination bulls whose semen was used at SFREC (fig. 3). To show how bull EPDs influence offspring performance in the absence of other effects and to contrast on-ranch and breed association EPDs, we projected possible offspring averages by resetting the EPD baseline to the SFREC herd average for weaning weight (540 pounds). If breed association EPDs were consistent with on-ranch progeny performance, all EPDs would be expected to fall on a diagonal line. However, this was not always the case. Some bulls with similar breed-association EPDs had significantly disparate on-ranch genetic evaluations, showing how on-ranch EPDs can help to distill actual genetic potential from the considerable variation associated with the low-accuracy EPDs typical of yearling bulls.
Fig. 3. Breed association expected progeny difference (EPD) for weaning weight, with calculated on-ranch weaning weight EPDs adjusted to represent potential observed weaning weights for both natural service (herd) and artificial insemination (AI) bulls.
Because the EPDs of heavily used artificial-insemination bulls already include progeny-test information, on-ranch evaluations do not offer an opportunity to greatly improve EPD accuracy. However, on-ranch evaluations can identify artificial-insemination sires whose offspring may be particularly well-suited to a given ranch environment. Using the integrated identification and tracking system, we tracked steer progeny of two high-accuracy artificial-insemination bulls from birth to harvest (fig. 4) (see box). Actual differences in performance average for birth weight, weaning weight and carcass weight were consistent with expected differences based on high-accuracy breed association EPDs for the same sires. In addition, this growth trend was consistent for two time points between weaning and harvest that are not typically included in breed-association genetic evaluations.
The use of integrated systems to develop high-accuracy EPDs for natural-service bulls could provide a powerful selection tool for commercial producers interested in improving their herds for feedlot and carcass traits. This will only occur if the market rewards producers for considering these traits in their selection criteria. The adoption of integrated identification systems depends on whether marketing systems provide monetary incentives for this information. Many cow-calf producers sell their calves at weaning and derive their income solely from the number and weight of calves sold. Therefore they never receive feedback regarding feedlot performance or information on how well those animals suited consumer preferences for tender, juicy meat. The fact that there is no financial incentive for cow-calf producers to consider many of these “downstream” traits in selection decisions effectively precludes genetic improvement for a number of important traits.
Fig. 4. American Angus Association (AAA) birth, weaning and carcass weight EPDs were similar to observed average values for steer progeny of two high-accuracy artificial insemination (AI) sires. Sire B's calves outperformed those of sire A from weaning through processing.
Tenderness evaluation
Increased tenderness has been associated with both consumer willingness to purchase and the price they are prepared to pay for beef (Platter et al. 2005). It is difficult to select for increased tenderness, a trait that can only be measured after the animal has left the herd. Traits that are difficult to measure, or are measured late in life, are well-suited to a DNA-based approach to estimate genetic merit (Allan and Smith 2008). Researchers are using molecular biology and quantitative genetics to identify regions of DNA that influence meat tenderness. DNA-marker tests developed to detect subtle sequence differences show whether a segment of an animal's DNA is positively or negatively associated with tenderness (Casas et al. 2006; Schenkel et al. 2006).
The two most prevalent marker tests for tenderness explain about 20% to 25% of the genetic variation for tenderness, and 12% to 18% of the overall variation in this trait (Van Eenennaam, Li, et al. 2007). These tests can be used for marker-assisted selection, which is the process of using the results of DNA-marker tests in a genetic-improvement program to assist in the selection of individuals to become the parents of the next generation.
As a field demonstration of the effect of these markers on meat tenderness, 40 genotyped steers from the 2008 calf crop at SFREC were selected and divided into two groups: one with the most tender genotypes, and the other with the least tender genotypes. Following harvest, meat samples will be collected from each carcass to take tenderness measurements. The Warner-Bratzler shear force (WBSF) protocol is the industry-accepted method of standardizing tenderness evaluations (Dikeman et al. 2005). It has three main components: (1) uniform handling and cooking of steaks, (2) core sampling steaks and (3) measuring the amount of force — equivalent to chewing — required to tear the core samples.
While tenderness is undoubtedly an important trait to consumers, it is less clear how selection for tenderness can provide an economic return to the cow-calf producer, who makes decisions based on factors for which they are paid — typically the number of calves sold and the price received for those calves. In the absence of a more integrated beef supply chain, it is likely that market failure will prevent tenderness from being an important consideration in breeding decisions on most commercial cow-calf ranches. However, some producers, such as Northern California's Prather Ranch, have developed vertically integrated niche markets for their beef, and are incorporating information from DNA tests for meat tenderness in their breeding decisions.
Whole-genome selection
Traditional genetic improvement of beef cattle relies on developing breeding values or EPDs for animals based on their performance and that of their relatives. Over the past 50 years, the selection of animals with the best breeding values has doubled the amount of milk that the average dairy cow produces in a year, and halved the amount of feed needed to produce a pound of pork. However, selection has not been as successful for traits that are difficult to measure, such as disease resistance, or traits that are not evident until late in an animal's life, such as fertility or longevity.
Technology breakthroughs developed during the sequencing of the human genome brought DNA-sequencing costs down, which made it economically feasible to sequence the genomes of other species. The bovine genome has recently been sequenced (Elsik et al. 2009), which has led to the discovery of many thousands of naturally occurring DNA-sequence variations in the form of SNPs between individuals (Bovine HapMap Consortium 2009). Researchers are now working to determine which variations are associated with desirable characteristics, such as disease resistance, in both humans and livestock species.
Information on variation in DNA sequences between animals may improve the accuracy of breeding values, that is, give breeders more confidence that they are selecting the best animals. Because DNA is available from birth, it may be possible to predict the genetic potential of animals at a very young age, in the absence of progeny testing, and keep only the best animals for breeding purposes. This may pave the way for producers to select animals to become parents of the next generation based on breeding values calculated from DNA-marker data alone, a process called “genomic selection” (Meuwissen et al. 2001). This approach may open the way to develop genetic predictions on difficult-to-measure traits, such as disease resistance and feed efficiency, that are not routinely included in beef-cattle genetic evaluations. It may also allow for the selection of traits that have never been previously considered in genetic evaluations, such as the compositional makeup and nutritional value of meat for human consumption.
Genomic technologies also offer new opportunities to develop management systems to optimize an animal's DNA genotype to best fit the production environment. For example, the genotype of some beef and dairy cattle may be better suited to grass-based production systems. It may also be possible to select animals that are able to grow to a given size using less feed, or that are more resistant to certain diseases. These technologies have great potential to enable the production of safer, more nutritious animal products. They may also allow for the selection of animals with a decreased environmental footprint and improved animal welfare due to lower levels of disease.
Farm advisor Dan Drake (right) and processing plant personnel examine the carcass of a UC steer. An electronic hand-held device loaded with appropriate software was used to capture the animal's electronic identification tag number, associate it with carcass attributes, and deliver that information to a database on the UC Davis server, where it was linked with earlier cow-calf and feedlot data.