All Issues
Plant and microbial research seeks biofuel production from lignocellulose
Publication Information
California Agriculture 63(4):178-184. https://doi.org/10.3733/ca.v063n04p178
Published October 01, 2009
PDF | Citation | Permissions
Abstract
A key strategy for biofuel production is making use of the chemical energy stored in plant cell walls. Cell walls are a strong meshwork of sugar chains and other polymers that encircle each plant cell. Collectively known as lignocellulose, cell wall material represents the bulk of plant dry mass. Biofuels can be made by releasing sugars from lignocellulose and converting them into fuel; however, this is currently an energy-intensive process. We summarize the barriers to efficient lignocellulosic biofuel production and highlight scientific research recently funded by the U.S. Department of Agriculture and U.S. Department of Energy, both to understand and harness the mechanisms by which plants build cell walls, and to further develop enzymes and microbes that facilitate sugar release and biofuel production.
Full text
Lignocellulosic biomass refers to vegetative tissues and plant-derived wastes that can be used to produce liquid transportation fuels. Postdoctoral Researcher Yuegeng Guan (left) and UC Berkeley professor Sheila McCormick of the Energy Biosciences Institute are studying Miscanthus sinensis at the USDA Plant Gene Expression Center in Albany.
Burning fossil fuels is inefficient and unsustainable, and it releases climate-changing carbon dioxide into the atmosphere. To ameliorate these problems, business leaders, policymakers and scientists are investigating alternatives such as producing liquid transportation fuels from plants. The production of ethanol and biodiesel fuels from food crops such as corn and soybeans is relatively energy-intensive and could potentially divert land from food production, leading to food price increases (Farrell et al. 2006). Still, these food-based fuels serve as a bridge to a future industry based on the use of vegetative tissues and plant-derived waste products, collectively known as lignocellulosic biomass (Waltz 2008).
Starch vs. lignocellulose
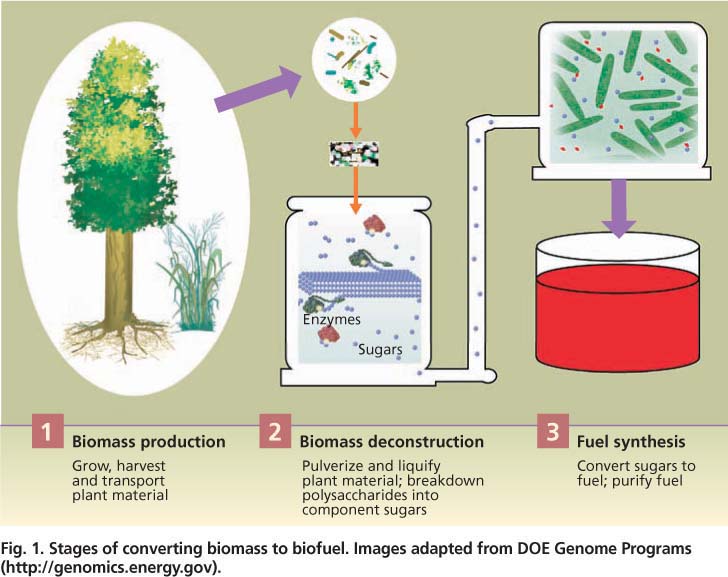
The conversion of both corn grain and biomass to fuels has the same three steps: (1) production of plant material, (2) deconstruction of the material into sugars and (3) conversion of the sugars into fuel (fig. 1). Corn-based ethanol production essentially follows the familiar process of brewing. First, starch is extracted from the corn grain and then cleaved into individual sugars with inexpensive enzymes. The resulting sugar, glucose, is fermented by yeast into ethanol.
Corn grain production, however, is highly inefficient compared with other diverse forms of biomass. Corn ears constitute only half of the aboveground tissue of the maize plant by dry weight, and only about 60% of the grain is starch (Somerville 2007). Thus, by utilizing only kernel-derived starch, more than half of the corn plant's sugar content is wasted. In contrast, lignocellulosic biomass is highly abundant, consisting of essentially the entire plant's dry mass, and including crop, forest and municipal wastes such as rice straw, wood chips and carbon-containing trash (Orts et al. 2008; Waltz 2008).
Additionally, analyses suggest that one of the most efficient and sustainable methods of biofuel production will be harvesting the aboveground portions of densely produced, fast-growing perennial energy crops such as poplar trees (Populus trichocarpa) and switchgrass (Panicum virgatum) (Schmer et al. 2008; Somerville 2007). The perennials under consideration require less water, fertilizer and management inputs compared with annual crops, and can be grown on marginal lands, including those with erosion-prone, dry or saline soils.
Fig. 1. Stages of converting biomass to biofuel. Images adapted from DOE Genome Programs ( http://genomics.energy.gov ).
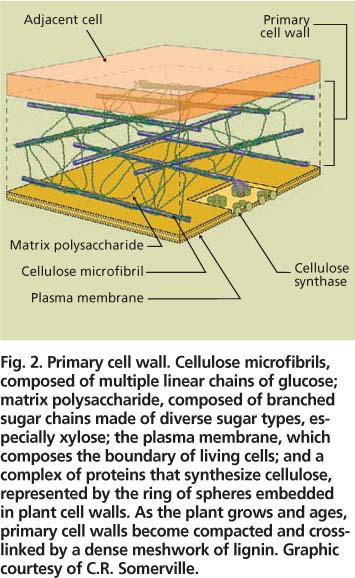
Fig. 2. Primary cell wall. Cellulose microfibrils, composed of multiple linear chains of glucose; matrix polysaccharide, composed of branched sugar chains made of diverse sugar types, especially xylose; the plasma membrane, which composes the boundary of living cells; and a complex of proteins that synthesize cellulose, represented by the ring of spheres embedded in plant cell walls. As the plant grows and ages, primary cell walls become compacted and cross-linked by a dense meshwork of lignin. Graphic courtesy of C.R. Somerville.
The potentially large energetic and environmental benefits of utilizing bioenergy crops and waste products — compared with fossil fuels and corn or other annual food crops — are greatly diminished by the current expense of deconstructing biomass into its component sugars (Lynd et al. 2008). Lignocellulosic sugar chains, called polysaccharides, are tightly bound to each other and are difficult to access. To release them, plant matter must be physically pulverized and then dissolved under energetically costly conditions, such as treatment with hot acid or pressurized alkali (Galbe and Zacchi 2007). Next, the polysaccharides are cleaved into individual sugars with enzyme catalysts, which are currently expensive, required in large amounts and do not cleave to completion (Galbe and Zacchi 2007; Lynd et al. 2008). As with current starch-based methods, the sugar water is then fed to yeast to produce ethanol. Yeast is currently unable to metabolize a significant fraction (as much as half) of the diverse sugars that compose cell walls.
Biofuel research goals
For each stage of production (fig. 1), researchers are pursuing technical solutions for the efficient use of lignocellulose to make biofuels, including integrating all steps in the process (Lynd et al. 2008). For research related to biomass production, major goals are to further reduce the environmental and monetary costs and increase the production efficiency of diverse bioenergy crops. High-density sources are necessary to reduce the costs of transporting biomass to processing stations. Another major goal is to understand and control how plants build cell walls in order to improve their deconstruction and synthesis into fuels.
For the deconstruction phase, major goals are to reduce the energy and other costs of pretreatment, as well as of the enzymes that catalyze sugar release. Fuel synthesis researchers are focusing on engineering organisms that can utilize 100% of the diverse sugars released from lignocellulose and tolerate the accumulation of high amounts of fuel product.
Due to the drawbacks of ethanol fuel, researchers are also exploring the synthesis of fuels less mixable (miscible) in water. These include butanol and alkanes, which may be used directly in conventional car engines. Beyond the scope of this review, researchers are seeking ways to utilize the nonsugar components of cell walls (Orts et al. 2008).
Cell wall synthesis
Plant cell walls consist primarily of two classes of polysaccharides — cellulose and diverse types of matrix polysaccharides (fig. 2). In addition, older, weight-bearing walls contain a network of the phenolic polymer, lignin.
Plants use their cell walls primarily for structural support, and gas and liquid transport; walls also participate in defense against fungi, insects and other pests and pathogens. Plant organs and cells with different functions have different cell wall compositions. The selective benefits of different plant taxa having distinct cell wall compositions are less clear (Carpita 1996). In particular, the matrix polysaccharides of grasses and other recently evolved monocots (plants with single seed leaves and parallel veination) are quite distinct from those of dicots (plants with two seed leaves and branched veins) (table 1) (Carpita 1996).
Researchers are investigating which genes are responsible for the synthesis and modification of plant cell walls and the biological function of each wall component. With this knowledge, they hope to manipulate plant walls to increase the efficiency of deconstruction, while still maintaining strong, vigorous plants that can be grown on a large scale. One idea being developed is to genetically engineer plants that express enzymes for breaking down cell wall components at the end of the growing season (Sticklen 2006).
Microfibrils.
Cellulose consists entirely of long, linear chains of the 6-carbon sugar, glucose. Glucose is the same sugar that makes up starch and, when depolymerized, is readily converted into ethanol by yeast. Cellulose typically composes nearly 50% of the cell wall (Pauly and Keegstra 2008). Bundles of 36 cellulose chains, called microfibrils, are laid down in a crisscross manner to form a scaffold — the cell wall equivalent of steel cables. Microfibrils are dense, crystalline structures that exclude water and become chemically cross-linked — and even less degradeable as plants grow and age. Large protein complexes at the cell surface synthesize microfibrils (fig. 2). Elusive for decades due to the difficulty of purifying active protein complexes from plant extracts, the plant genes for cellulose synthesis were finally identified in 1996, based on sequence similarity to bacterial cellulose synthases (Pear et al. 1996). An important remaining question is how plants regulate the synthesis of cellulose to integrate microfibril synthesis with other wall components.
Matrix polysaccharides.
Between the microfibrils, shorter, branched matrix polysaccharide chains interlace (fig. 2). Matrix polysaccharides are enriched for the 5-carbon sugar, xylose, and compose about 30% of the cell wall (Pauly and Keegstra 2008). The grass matrix consists primarily of mixed polysaccharides of the sugars xylose and arabinose (Carpita 1996); whereas, the matrix of dicots, like poplar and alfalfa, are mostly polysaccharides of glucose, xylose and glucuronic acid as well as diverse and complex pectin polysaccharides.
Matrix polysaccharide is more easily extracted from the cell wall than cellulose; however, most yeast cannot use the 5-carbon sugars that make up the bulk of the matrix. Moreover, matrix components often contain chemical modifications that inhibit the breakdown of cellulose and other downstream processes. While genes for the synthesis of many of the major matrix components of dicots have been defined (Farrokhi et al. 2006), most of the genes for grass-specific matrix synthases and modifying enzymes have not yet been assigned and characterized. Strategies for identifying the genes responsible for the synthesis of cell wall components include examining the results of introducing the putative cell wall–synthesis genes into living systems that normally lack the genes, and disrupting or increasing the expression of such genes in their plant hosts (Farrokhi et al. 2006). Screening plant populations for natural or induced genetic variation in cell wall structure and function is also a common approach (Brown et al. 2005).
Lignin.
As plants grow and require stronger walls, lignin is deposited in the cell wall (15% to 30% in mature walls) (Boerjan et al. 2003; Pauly and Keegstra 2008). Individual lignin monomers — the main portion of which is a carbon ring — react with various wall components, particularly other lignin, to cross-link and reinforce the cell wall. The result is a chemical meshwork that is physically strong and difficult to degrade. Thus, decreasing the strength of the lignin network is a prime target for genetic manipulation in plants (Chen and Dixon 2007).
For example, Clint Chapple of Purdue University and colleagues are analyzing the effect of blocking lignin-synthesis genes on cell wall composition and growth in poplar (Coleman et al. 2008). Also, John Ralph and associates at the University of Wisconsin have shown that plant cells can be fed modified lignin monomers (simple compounds), with the result that the lignin they then produce has chemical “zippers” and can be more easily deconstructed or “unzipped” (Grabber et al. 2008). If methods can be devised to develop plants that make such modified lignin, the plants' cell walls might be more easily deconstructed while maintaining their biological functions. Researchers are pursuing this goal at the Joint BioEnergy Institute (JBEI; www.jbei.org ) in the San Francisco Bay Area and the Great Lakes Bioenergy Research Center ( www.greatlakesbioenergy.org ), both with funding from the U.S. Department of Energy (DOE).
Jay Keasling (left) and Rajat Sapra of the Joint BioEnergy Institute developed a technique that speeds up the search for improved microbes to ferment plant sugars into biofuels.
Reference plants
A major research strategy to elucidate the synthesis and breakdown of cell wall components is to conduct experiments with well-characterized reference plants. The results can then be applied toward improving species that produce significant amounts of biomass (table 1). The best-characterized plant at the molecular level is a diminutive species from the mustard family, Arabidopsis thaliana, which has a short generation time of 6 to 8 weeks. The research community has developed abundant resources and methods to support examination of Arabidopsis gene functions. Arabidopsis was the first plant to have its DNA fully sequenced (AGI 2000). This has allowed scientists to identify Arabidopsis plants with mutations in almost every gene, so that the effect of deleting most genes of interest can be rapidly examined (Alonso et al. 2003). Currently, the U.S. National Science Foundation is supporting the collection of significant functional information for every Arabidopsis gene. The basic knowledge gained from gene function studies in Arabidopsis can often be applied to other plant species, especially broadleaved dicots such as poplar (table 1).
In addition to Arabidopsis, the grass-family member rice (Oryza sativa) has also been developed as a reference plant. It also has short generation times (9 weeks for some varieties) and is easily transformed (Jung et al. 2008). Grasses share a high degree of similarity in the arrangement and, in many cases, the function of their genes (Devos 2005). This suggests that rice data will greatly assist with understanding other grasses, including the many being developed as energy crops in the United States (table 1). The rice genome, smallest among the cereals, has been fully sequenced, and rice mutant collections are being developed to facilitate gene function studies (Goff et al. 2002; Krishnan et al. 2009). Genes that have high sequence similarity between Arabidopsis and rice are generally expected to function in a similar, though not identical, manner. However, only 50% of rice genes are closely related by sequence similarity to an Arabidopsis gene (Goff et al. 2002). For example, grasses possess distinct groups of genes likely involved in cell wall synthesis that are not represented in the genomes of Arabidopsis and other dicot plants (Cao et al. 2008).
In an example of model plant studies, JBEI researchers are determining how particular genes contribute to the cell wall structures in rice and Arabidopsis. Their strategy includes isolating enzymes involved in cell wall synthesis from rice and Arabidopsis to determine their biochemical effects on purified sugars (Jensen et al. 2008). The results of these studies will provide information relevant to the two major classes of flowering-plant cell walls, those of dicots and grasses (table 1).
Genomic data
The identification of part or all of the genetic sequence of a species provides invaluable information for improving yield and other characteristics. The DOE's Joint Genome Institute, located in Walnut Creek, Calif., has collaborated with other researchers to sequence the genomes of the potential energy crops sorghum (Sorghum bicolor) and poplar (Paterson et al. 2009; Tuskan et al. 2006). Given the expected importance of grasses as biomass producers, DOE is also sequencing the genome of the small, dryland grass Brachypodium distachyon, which is being developed as a reference species like rice and Arabidopsis (Opanowicz et al. 2008). A complementary approach to sequencing the full genome is to sequence just the expressed genes (messenger RNA) isolated from different tissues. The U.S. Department of Agriculture (USDA) and DOE have sequenced about 500,000 segments of expressed genes from switchgrass (Tobias et al. 2008), and progress is now under way to sequence its complete genome.
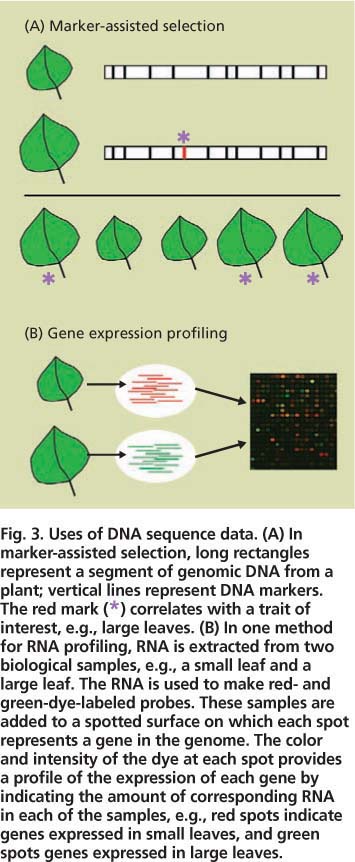
Fig. 3. Uses of DNA sequence data. (A) In marker-assisted selection, long rectangles represent a segment of genomic DNA from a plant; vertical lines represent DNA markers. The red mark (*) correlates with a trait of interest, e.g., large leaves. (B) In one method for RNA profiling, RNA is extracted from two biological samples, e.g., a small leaf and a large leaf. The RNA is used to make red- and green-dye-labeled probes. These samples are added to a spotted surface on which each spot represents a gene in the genome. The color and intensity of the dye at each spot provides a profile of the expression of each gene by indicating the amount of corresponding RNA in each of the samples, e.g., red spots indicate genes expressed in small leaves, and green spots genes expressed in large leaves.
The uses of sequence data are many. We highlight just two techniques that researchers are using for energy crops, marker-assisted selection and biomolecule profiling analysis (fig. 3).
Marker-assisted selection.
Marker-assisted selection accelerates the development of improved plant varieties by taking advantage of DNA sequences that vary slightly among different plants in a population (fig. 3) (Robins et al. 2007). Each sequence variant is known as a genetic “marker.” Plant geneticists examine a particular trait of interest, such as large leaves, for association with DNA markers. In the ideal case, a DNA marker (or markers) might be found to occur in every plant that has the trait but none of the plants that lack it. Thus, breeders can quickly screen large numbers of plants with a simple assay for the marker rather than conduct more time-consuming tests for the actual trait, for example by manually measuring leaves at a particular growth stage.
In recently funded work, scientists initiated a number of marker-assisted selection programs for potential bioenergy crops. Researchers led by Edward Buckler at Cornell University and Christian Tobias at the USDA Western Regional Research Center in Albany, Calif., are looking for markers associated with biomass production traits, such as plant height, tiller number and photosynthetic rate in switchgrass and reed canary grass. William Rooney of Texas A&M University and E. Charles Brummer of University of Georgia are conducting similar studies in sorghum and alfalfa, respectively (Murray et al. 2009; Robins et al. 2007). These researchers will also be looking for markers associated with the quality of the biomass for biofuel production, in terms of sugar availability. Because many dedicated bioenergy-crop species have not previously been the subject of intensive breeding efforts, researchers expect that significant improvements can be made relatively quickly.
Profiling analysis.
Another application of sequence data is to develop large-scale profiling approaches to simultaneously measure the participation of many gene products in a biological process (fig. 3) (Jung et al. 2008). For example, cell wall synthesis is carried out by only a subset of plant proteins. At the appropriate time and plant tissue, the cell wall genes are read out (expressed) as messenger RNAs that are then translated into proteins, such as the enzymes that synthesize cellulose and other polysaccharides. Profiling approaches monitor which RNAs or proteins are present in a particular biological sample such as an expanding poplar leaf. Such approaches require sequence data because usually only a small fragment, or tag, of each RNA or protein is revealed during profiling, and sequence data allows this tag to be equated with the full-length gene (fig. 3).
A recently funded consortium project involving University of Mississippi, UC Davis and The Ohio State University entails RNA and protein profiling of rice cells as they regenerate their stripped-away cell walls. Having identified potentially important genes for cell wall synthesis via profiling, these researchers are characterizing mutant plants in which the identified genes have been disrupted, in contrast to other mutants in which the identified genes have increased levels of expression (Brown et al. 2005).
Cell wall deconstruction
Microbiology research to support biofuel production primarily addresses the deconstruction and fuel synthesis stages of biofuel production (fig. 1). JBEI seeks to help lead the development of improved enzymes for the more effective breakdown of lignocellulose. That consortium is focusing on enzymes to break down lignin, the cross-linking polymer that greatly reduces the availability of cellulose in mature plant tissue. The JBEI strategy begins by developing improved assays for monitoring the cleavage of various cell wall components simultaneously. These improved measurement methods will allow screening of potential sources of new lignocellulose-degrading enzymes from environments where cell wall degradation occurs, such as compost heaps and rainforest floors.
Samples that provide effective cleavage of key wall components can be analyzed by DNA sequencing, as, for example, has recently been performed on the community of organisms inhabiting the hindgut (stomach) of a termite species (Warnecke et al. 2007). Such sequencing of whole communities, as opposed to single organisms, is known as metagenomics. With such an approach, deconstruction enzymes can be identified without the requirement to be able to isolate and grow the organism that makes them. Full genome sequencing of important organisms from lignocellulose-degrading environments is another strategy for identifying genes for deconstruction. In this vein, researchers have recently sequenced the genome of a key cellulose-degrading bacterium, again from the guts of termites (Hongoh et al. 2008).
Seeking an alternative to yeast, Veronica Fok of the Joint BioEnergy Institute in Emeryville is engineering new microbes that can quickly and efficiently ferment complex sugars into advanced biofuels.
Newly discovered and formerly characterized enzymes will be further studied and subjected to directed evolution to develop improved enzymes that are more effective under commercial conditions. Directed evolution generates new variants of the enzymes of interest through repeated cycles of mutation and selection. Variation can be introduced into the enzyme target by numerous methods, including gene shuffling, which results in replacement of pieces of the enzyme with pieces from other similar enzymes (Shibuya et al. 2000). The new enzyme variants are then screened for improvements in the desired activity. Any variants that pass the screen can then be fed back through the cycle of mutation and further screened with increased stringency or different criteria.
Conversion of sugars to fuel
The currently employed, yeast-based method of sugar conversion to ethanol has a number of shortcomings for fuel synthesis. The fermentation of sugars by yeast does not proceed to completion nor does it utilize two of the major carbon sources in lignocellulose — the 5-carbon sugars that are abundant in matrix polysaccharides and phenolic lignin. Furthermore, ethanol as a fuel is not optimal; it has low energy content (67% less than gasoline), requires energy to separate from water and is corrosive (Somerville 2007). Therefore, the major goal for biofuel production is to develop an organism or community of organisms that utilizes all of the major components of lignocellulose and produces a more gasoline-like fuel. Ideally, this organism(s) would also produce cell wall–degrading enzymes, incorporating many of the deconstruction functions of cleaving polysaccharides and lignin networks in the fuel synthesis phase.
Many of the biochemical processes required for the conversion of lignocellulose into diverse fuels have already been identified in various organisms, though the search for improved or alternative chemistries continues. Engineering or selecting for high yields is a crucial area of research. Another major challenge is to bring together all of the processes in a single organism, or a few coordinated organisms, that perform well under industrial conditions. For example, some anaerobic bacteria can ferment 5-carbon sugars. Based on the pathways in those organisms, laboratory yeast strains that are intolerant of industrial conditions have been engineered with this ability (Jeffries and Jin 2004). Other organisms have been described that produce alternative fuel products. For example, the bacterium Vibrio furnissii converts glucose to alkanes, long hydrocarbon chains similar to those found in petroleum (Park 2005). In a recent elegant example of combined metabolic engineering and directed evolution, the commonly used bacterium Escherichia coli has been engineered to produce relatively high yields of butanol (Atsumi et al. 2008). Butanol is a 4-carbon alcohol that can be directly substituted for gasoline in unmodified car engines, though it still lacks beneficial characteristics compared with less water-miscible alkanes such as hexadecane.
JBEI and other groups will be working on moving the necessary enzymes into yeast and other organisms for use on an industrial scale. Using such techniques as RNA, protein and small-molecule profiling similar to those described here for examining plants, the biological state of the engineered organisms will be monitored to evaluate how to make improvements. These studies will build on progress in understanding microorganism biology, which has already advanced to using mathematical models that accurately describe the flow of sugars and small molecules through the series of enzymes that convert sugars to fuels (Suthers et al. 2007).
The biofuel future
Significant creativity is now being focused on the challenge of developing sustainable biofuel production. Optimizing the efficiency of lignocellulose breakdown is a major goal. Researchers also seek to improve bioenergy crop production and develop improved fuel synthesis methods. With such advances, biofuel production has the potential to ft into a near- to midterm future in which transportation fuels are efficiently generated with little greenhouse-gas production from municipal and agricultural waste products and low-input, extremely high-yielding perennial energy crops. The current DOE goal is that by 2030, 30% of U.S. transportation fuels will come from from such alternative sources (Perlack et al. 2005).