All Issues
PCR and antibody methods: Research compares two cattle feed tests that detect bovine byproduct contaminants
Publication Information
California Agriculture 59(4):212-226. https://doi.org/10.3733/ca.v059n04p212
Published October 01, 2005
PDF | Citation | Permissions
Abstract
Preventing the spread of mad cow disease through contaminated cattle feed is a major concern of beef and dairy producers, regulators and consumers around the world. Routine testing of cattle feeds for the presence of banned substances is a critical control point in assuring animal health and food safety. We compared the results of two test procedures (a real-time polymerase chain reaction [PCR] assay and a commercially available ruminant antibody detection kit) on five cattle rations spiked with bovine meat-and-bone meal, or with bovine dried blood. The real-time PCR consistently detected these contaminants at lower levels in each of these diverse cattle rations.
Full text
Bovine spongiform encephalopathy (BSE), also known as mad cow disease, has now been found in 26 countries including Canada and the United States. The consumption of meat from BSE-infected cattle is believed to have caused the deaths of close to 200 people worldwide, from a disease called variant Creutzfeldt-Jakob disease (vCJD) (GAO 2005). Frthermore, BSE can have devastating effects on a country's beef industry. More than 5 million cattle were killed in an effort to control BSE in Europe; in the United Kingdom alone, almost 4 million head of cattle were destroyed through January 2004, costing the British economy as much as 5 billion pounds (the equivalent of up to 8 billion dollars).
When a single BSE-infected cow was found in the United States on Dec. 23, 2003, major foreign markets prohibited the entry of U.S. cattle. The immediate effect was that the beef industry lost more than 80% of its export trade, or an estimated $2 billion dollars. A second U.S. case was confirmed on June 24, 2005, leading to extensions of the feed bans. Fifty-nine countries have now imposed import bans or restrictions on American beef, virtually wiping out a $3 billion export market ( see page 203 ).
Gerald Johnson, manager of the UC Davis feed mill, advises researchers and supplies them with custom-milled feeds required for their projects. The feed mill, built in 1959 by the California Feeder Council and numerous other donors, is still in daily use.
The transmission of BSE in cattle most probably occurs through the ingestion of feed that contains rendered byproducts of BSE-infected ruminants (Wilesmith et al. 1988). In 1997, the United States banned the use of “most mammalian proteins” in the manufacture of feed for cattle and other ruminants (Federal Register 1997).
Detecting the presence of illicit feed additives is critical; risk assessors believe that if there were full compliance with the ban, a U.S. case of BSE would be self-limiting (HCRA 2003). However, the diagnosis of BSE in a third cow in Canada (Jan. 11, 2005) points to a weak link in the system: compliance. Investigation confirmed that the affected Canadian animal was fed contaminated feed that had been produced before the ban was established, illustrating both the consequences of noncompliance with the illicit additive ban (the rancher used this contaminated feed after the ban) (GAO 2005), and that contaminated feed may still exist in Canada (Skelton 2004). A $7 billion class-action suit representing 100,000 Canadian farmers accuses the Canadian government of negligently allowing mad cow disease to devastate their cattle industry (Makin 2005).
At present, the FDA accepts only one method of testing ruminant feed for banned substances, a microscopic examination for the presence of animal tissue such as hair and bone particles (GAO 2005). However, other tests are under development for FDA consideration. At present, the FDA and state regulatory agencies use the latter tests for initial screening but not confirmation. We performed a side-by-side comparison of two of these detection methods, evaluating their ability to detect ruminant contamination in cattle feeds. The tests were distinctly different: a real-time polymerase chain reaction (PCR)-based assay (developed by our laboratory) that detects ruminant-specific DNA, and a commercially available, antibody-based, latera flow assay that detects ruminant muscle protein (Reveal, Neogen, Lansing, Mich., Product #8100, lot 16096B).
UC Davis veterinarian Jim Cullor heads the team that is developing tests for animal contamination in livestock feeds.
DNA and antibody-based assays
The need to detect DNA at the lowest possible levels led to the early application of PCR technology for bovine DNA (Tartaglia et al. 1998; Wang et al. 2000; Kremar and Rencova 2001). The PCR method we have developed relies on species-specific variation of mitochondrial DNA (mtDNA). MtDNA has two features that make it appealing as a species-specific marker. First, it is abundant, present at 1,000 times higher concentrations than most single-copy nuclear genes, allowing for an extremely low limit of detection. Second, because mtDNA evolves much more rapidly than nuclear genes, there is substantial species-specific variability upon which to design species-specific PCR reagents, those that bind to the mtDNA of a particular species. An additional advantage of using PCR assays to test cattle feeds is that DNA is heat-stable and may be detected even in rendered products, which have been subjected to high-heat processing (244°F to 289°F). In contrast, proteins can be substantially denatured by such high heat and made unrecognizable to antibody-based assays.
In current practice, feed is tested by both the FDA and state officials who have contracts with FDA to test and inspect animal feed. Samples of feed are taken for testing from manufacturing facilities, bulk feed sold to cattle feedlots and bags of feed sold at retail stores. No amount of any banned substance is allowed, regardless of how small the concentration. In the event that prohibited materials are found in the feed, the FDA can either issue a warning letter, ask firms to voluntarily issue a recall of the feed, or get a court order to seize the feed and feed ingredients (GAO 2005).
In the PCR analysis method we developed, a feed sample is first digested chemically to release nucleic acids. We then multiply (amplify) these nucleic acids, using PCR technology. Additional PCR reagents (probes) are used to detect the PCR products. This analysis takes advantage of several advanced technologies, including real-time fluorescent instruments that offer extremely fast real-time monitoring of PCR amplification reactions. In addition, we can achieve further specificity of the result via a melting-curve analysis in which the double-stranded PCR product is melted apart. Since this melting temperature value is directly related to the DNA sequence of the amplified product, it provides a fast, more reliable and less labor-intensive way to verify the identity of the amplified product in every sample assayed. The use of fluorescent resonance energy transfer (FRET) probes further increases the lower limit of detection, making the assay even more sensitive than conventional PCR methods. These innovations have greatly improved laboratory sensitivity for the detection of specified risk materials in feed (Rensen et al. 2005). While sensitivity levels vary, in our trials the PCR method detected the presence of contaminants when the other tests did not. However, while PCR technology is sensitive and accurate compared with other methods, the equipment to run such tests is expensive and requires laboratory support.
The commercial, antibody-based, lateral-flow kit — called Reveal — addresses the need for a field assay for mammalian proteins. This method uses antibodies on a test strip to detect the presence of ruminant proteins in the sample. When the antibodies bind to ruminant proteins, the test strip develops a colored band within 15 minutes. While typically not as sensitive as DNA-based assays, antibody-based assays are much simpler and easier to perform. Producers could use such field tests at their livestock facilities.
Testing different feed types
PCR test
To test the efficacy of these two assays under laboratory conditions, we spiked five representative types of cattle feed down to 0.1% w/w concentrations of either bovine meat-and-bone meal (BMBM) or bovine dried blood (BDB). These low concentrations may represent accidental contamination of feed during processing. Prior to real-time PCR analysis of the feed samples, DNA extraction was accomplished using modifications of a commercial kit, adapting the protocol to accommodate a larger sample size (0.22 gram) (Qiagen Plant Kit, Qiagen Inc, Valencia, Calif.). Detection and analysis were performed on each concentration of BMBM and BDB through fluorescent real-time PCR using the LightCycler (Roche Applied Sciences, Indianapolis, Ind.) (Rensen et al. 2005).
Before applying PCR analysis, each feed sample was ground to a fine powder and BMBM or BDB was added at the specified concentration. RNAse (DNA- and RNA-free) (Roche Applied Sciences, Indianapolis, Ind.) was added at a rate adjusted to the volume of the shredder column nucleic acid eluate (Sawyer 2004). After the final extraction, the concentrated DNA was aliquotted and subjected to real-time PCR analysis.
Antibody-based test
Using the antibody-based Reveal test kit analysis, the five cattle feeds were processed according to the kit instructions. The appropriate spiking amount of BMBM or BDB was added directly to the extraction vessel to attain a total of 10 grams of spiked feed. After swirling and then boiling for 10 minutes, the sample was removed and swirled again. An aliquot of the digested liquid was immediately transferred to a microcentrifuge tube. A test strip was inserted and evaluated after precisely 15 minutes. The test is considered positive when colored bands develop in both the control and target zones. Unspiked feeds were included as negative controls.
Feed types
The type of feed has been reported to be an important factor in how well these two tests detect ruminant protein or DNA (Sawyer et al. 2004). We tested five cattle feeds with a range of concentrate-to-roughage ratios: (1) finishing ration, 80-to-20, without molasses or bovine tallow; (2) finishing ration, 80-to-20; (3) starter calf ration, 40-to-60; (4) grower calf ration, 60-to-40; and (5) weaning calf ration, 70-to-30 (Trophy Maker Calf Maker, Alderman-Cave Milling and Grain Company of New Mexico, Roswell, N.M.). The first feed was the only type tested in which molasses and bovine tallow were originally excluded due to anticipated sample processing problems. All the other feeds tested included 0.01% to 0.04% molasses and 1.5% to 2.5% bovine tallow, as determined by the formulations.
Confounding factors
Roughage concentration
RNA and other inhibitors released from components of feed during sample digestion have been implicated in false-negative PCR results, where chemicals in the sample may interfere with the enzymes used in the PCR reaction. Treating the nucleic acid extract with RNAse (enzymes that break down RNA) prior to PCR results in a consistently lower DNA limit of detection (Sawyer et al. 2004). In a previous study, feeds containing the highest amounts of roughage were most frequently associated with false-negative PCR results. We were consistently unable to achieve the same lower limit of detection obtained with lower roughage feeds (nos. 1 and 2, 20% roughage) when analyzing higher roughage feeds (nos. 3 and 4, 60% and 40% roughage, respectively).
In the current study, the melting-curve analysis indicated that 0.01% BMBM in the two higher-roughage feeds was detected near the threshold limit of detection (fig. 1). With the antibody-based Reveal assay, the highest roughage feed (no. 3) produced inconclusive results at the same concentration of BMBM (table 1). This supports the theory that roughage may be an inhibitory factor in both the DNA- and antibody-based assays for detecting ruminant contamination of cattle feed.
Detecting blood
The antibody-based Reveal test was unable to detect blood at all since the antibody only binds to a muscle protein (troponin). As for the DNA-based assay, the bovine-specific mtDNA primers used can only detect nucleated cells. Most of the BDB cells are mature red blood cells, which are nonnucleated. White blood cells (which are nucleated) contribute only about 1% of the total cellular component of dried blood (Kramer 2000). In contrast, BMBM contains much greater numbers of nucleated cells than BDB and thus has a greater probability of being detected by the DNA-based assay. Both BMBM and BDB are commercially available rendered products. When testing for these combined products in cattle feed, the lowest limit of detection depends on the product or product combination, the type of ration being tested and their respective concentrations in the sample. BDB would require higher concentrations as it is the least detectable tissue.
Comparison of assays
Limits of detection
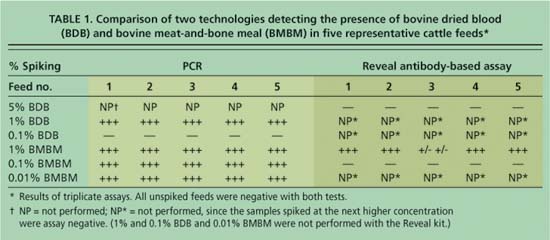
The DNA-based real-time PCR technology consistently detected BMBM in three replicate samples of all five feeds at the 1%, 0.1% and 0.01% limits of detection. BDB was detected at the 1% but not at the 0.1% limit of detection (table 1).
The antibody-based Reveal test, also performed in triplicate, detected BMBM at the 1% limit of detection in all feeds except no. 3, which was inconclusive. However, the Reveal assay did not detect BMBM in any of the feeds at the 0.1% limit of detection. While the presence of 5% BDB was readily observed visually in the digested feed sample fluids (see picture); as anticipated, the Reveal test strip did not detect BDB at this concentration since the specific antibody to troponin, the muscle protein, does not cross-react with blood cells. Failure to detect BMBM at the 1% level of spiking in some feed types or to detect BDB at any level of spiking appears to be a disadvantage of the Reveal assay.
Fig 1. A permanent record of results is provided by the printout of the melting-curve analysis of amplified PCR products. The five feeds spiked with 0.01% bovine meat-and-bone meal (BMBM) exhibit characteristic melting-temperature peaks of 143.6°F-145.4°F (62°C-63°C), similar to the positive control (tall, red peak). The negative PCR system control (blue, baseline peak) demonstrates the absence of any aberrant reagent contamination. The other five peaks identify product in each of the five feeds as BMBM.
TABLE 1. Comparison of two technologies detecting the presence of bovine dried blood (BDB) and bovine meat-and-bone meal (BMBM) in five representative cattle feeds*
Subjectivity
The LightCycler melting-curve analysis of results printout — provided by the PCR assay — provides a permanent record for litigation or enforcement documentation (fig. 1).
On the other hand, in these trials the results of the Reveal test at the lower (1%) limit of detection were subjective and ambiguous. In each case, a defined positive control line was apparent within 5 minutes, however most of the test samples required 15 minutes to develop a barely perceptible line. For some samples, the intensity of the test sample line increased and became more apparent with an additional 10 to 15 minutes, but in all cases never attained the intensity of the positive test line (see picture, page 216). The subjectivity in interpretation of these test-strip results and the fact that the test sample line became more apparent over time suggests that it may not be possible to use the stored test-strip as an accurate and permanent record.
Time
The Reveal test was developed for field use. It offers the obvious advantages of on-site convenience, flexibility and time savings. In contrast, the PCR technology test requires laboratory support, augmented by a sophisticated analysis system that is dependent upon processing large numbers of samples in order to be cost-effective in an agricultural setting.
The real-time PCR process has three phases: 1.25 hours for DNA preparation and extraction (including sample grinding and weighing); 0.5-hour to set up the real-time PCR reaction; and 0.75 hour to 2.5 hours per sample for the real-time PCR analysis. This adds up to 2.5 to 4.25 hours per sample. However, our current real-time fluorescent PCR systems are able to assay from 32 to 96 samples simultaneously, which translates to 512 to 1,536 samples per 40-hour workweek.
In contrast, Neogen states that the total Reveal sample-processing time is less than 0.5 hour. With duplicate equipment, one person may be able to conveniently process five samples simultaneously in that time-span, which translates to 400 samples per 40-hour workweek. Taking advantage of batch processing, real-time PCR technology could process about 1.3 to 3.8 times as many samples as Reveal in a 40-hour workweek.
Cost
The direct costs per sample for both technologies, including supplies and technical help, are comparable ($8.87 to $9.47 per sample for real-time PCR, and $9.20 per sample for Reveal). (The initial cost of real-time PCR equipment, about $50,000, would be borne by a laboratory.)
Despite the obvious disadvantage of not being a field test, real-time PCR offers a considerably lower limit of detection and greater accuracy. The indirect costs of the antibody test could be large if the result was a false negative in the field that was ultimately confirmed positive in the laboratory (Wyatt 1992), even if the contaminated feed did not result in a case of BSE. Moreover, the loss of public confidence is less quantifiable but equally significant.
Comparison of the visual appearance of tubes containing test fluid of feed sample no. 5, spiked to attain 5% bovine dried blood (BDB) (left) and an unspiked sample (right). The presence of BDB in fluid to be tested by the Reveal assay is readily identifiable on the left. Blood cells are floating at the interphase (arrow) and suspended in the fluid and sediment (arrow). Since the antibody incorporated in the Reveal test strips does not cross-react with blood cells, the same sample (5% BDB) tested negative. Samples of all five feeds spiked to attain only 1% BDB tested positive with the realtime PCR technology, which detects the bovine mitochondrial DNA in nucleated white blood cells.
Sampling errors
In contrast to the DNA-based assay, the antibody-based Reveal assay can accommodate a larger sample size and the sample does not have to be finely ground prior to processing. In general, a larger-size sample will increase the ability to detect contamination at lower concentrations.
Grinding the feed sample to a powder, as required for the PCR test, does yield a denser, more consistent sample, and thorough mixing of the ground, spiked sample is essential for homogeneously dispersing the contaminating BMBM or BDB prior to weighing the aliquots for testing. As the instructions in the Reveal test kit point out, the particulate contaminant could easily sift out through the fibers of the feed and be missed when the sample is collected. Likewise, in practice the contaminant would tend to sift out through the feeds being tested. Because these contaminants are granular substances, great care must be taken at all stages of the process to obtain a sample that contains particulate material, regardless of the kind of assay chosen.
Reveal test strips performed on feed no. 5 are displayed, top to bottom: unspiked negative control; feed spiked to attain 1% bovine meat-and-bone meal (BMBM); 0.1% BMBM; and 5% bovine dried blood (BDB). All positive control bands are clearly seen as the red lines in the middle of the strip. The faint sample band line observed in the 1% BMBM sample (arrow) was considered positive at the development time of 10 minutes. A positive sample band failed to develop with the 0.1% BMBM or 5% BDB spiked samples.
Advantages and disadvantages
The ability to prevent ruminant byproduct material from entering ruminant feed is essential to preventing BSE transmission. A method to detect these contaminating byproducts is necessary, not only to detect and deter the intentional illegal amending of feeds, but also to detect inadvertent contamination due to inadequate clean-out procedures, material mislabeling, or other production errors. Because the results of these tests for ruminant proteins could lead to legal action, it is important that they be objective, definitive and reproducible. Additionally, results should provide a permanent record.
Both assays offer advantages and disadvantages, and provide tools for producers, processors and regulators in particular settings. In these trials, consistent antibody-based Reveal results were obtained with four of the five feeds at a concentration of 1% BMBM; however, the inconclusive result with the 1% BMBM-spiked sample of feed no. 3 suggests that a negative Reveal test should not be considered reliable with certain feeds in the lower concentrations (1% and below) (table 1). The ability to detect 1% BDB via the DNA-based real-time PCR is of potential importance. At the time of this publication, blood had been removed from the FDA list of banned ruminant feeds due to the lack of evidence that it transmits BSE, but it is still considered a material of concern and may be added back to the list in the future. We conclude that despite the disadvantages of time, convenience and cost, the consistent detection of smaller amounts of contamination is more likely with the more sensitive, quantitative, real-time PCR analysis.